Wrapping up
Loose ends and more content than we’ll get to
Daniel Anderson
Week 10
Agenda
Review Homework 3
RQ to models practice
Finishing up the last bits from last time
Quick discussion on missing data
Cross-classified models
- And a brief foray into multiple membership models
Bonus info for you if you want to cover on your own
- Fitting a 1PL model through a mixed effects framework
Data
alc## # A tibble: 246 x 6## id age coa male alcuse peer## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 14 1 0 1.732051 1.264911 ## 2 1 15 1 0 2 1.264911 ## 3 1 16 1 0 2 1.264911 ## 4 2 14 1 1 0 0.8944272## 5 2 15 1 1 0 0.8944272## 6 2 16 1 1 1 0.8944272## 7 3 14 1 1 1 0.8944272## 8 3 15 1 1 2 0.8944272## 9 3 16 1 1 3.316625 0.8944272## 10 4 14 1 1 0 1.788854 ## # … with 236 more rowsHow I would start
First, estimate the model that makes the most theoretical sense, to me - random intercepts and slopes
If that model fits fine, just go forward with it and interpret
If that model does not fit well (i.e., convergence warnings), contrast that model with one that constrains the slope to be constant across partcipants
Model summary
summary(rq1a)## Linear mixed model fit by REML ['lmerMod']## Formula: alcuse ~ age_c + (1 + age_c | id)## Data: alc## ## REML criterion at convergence: 643.2## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -2.48287 -0.37933 -0.07858 0.38876 2.49284 ## ## Random effects:## Groups Name Variance Std.Dev. Corr ## id (Intercept) 0.6355 0.7972 ## age_c 0.1552 0.3939 -0.23## Residual 0.3373 0.5808 ## Number of obs: 246, groups: id, 82## ## Fixed effects:## Estimate Std. Error t value## (Intercept) 0.65130 0.10573 6.160## age_c 0.27065 0.06284 4.307## ## Correlation of Fixed Effects:## (Intr)## age_c -0.441RQ 3
02:00
Do male children of alcoholics have higher alcohol use than female children of alcoholics?
Important - we're specifically interested in coa == 1 cases.
However: We have to model main effects for coa and male to get at male:coa.
rq3 <- lmer(alcuse ~ age_c + coa + male + coa:male + (1 + age_c | id), data = alc)arm::display(rq5)## lmer(formula = alcuse ~ age_c + coa + male + age_c:male + age_c:coa + ## coa:male + age_c:male:coa + (1 + age_c | id), data = alc)## coef.est coef.se## (Intercept) 0.42 0.19 ## age_c 0.22 0.12 ## coa 0.76 0.29 ## male -0.22 0.27 ## age_c:male 0.15 0.17 ## age_c:coa -0.23 0.18 ## coa:male 0.00 0.40 ## age_c:coa:male 0.32 0.25 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 0.72 ## age_c 0.37 -0.20 ## Residual 0.58 ## ---## number of obs: 246, groups: id, 82## AIC = 653.4, DIC = 597.1## deviance = 613.3arm::display(rq5b)## lmer(formula = alcuse ~ age_c * coa * male + (1 + age_c | id), ## data = alc)## coef.est coef.se## (Intercept) 0.42 0.19 ## age_c 0.22 0.12 ## coa 0.76 0.29 ## male -0.22 0.27 ## age_c:coa -0.23 0.18 ## age_c:male 0.15 0.17 ## coa:male 0.00 0.40 ## age_c:coa:male 0.32 0.25 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 0.72 ## age_c 0.37 -0.20 ## Residual 0.58 ## ---## number of obs: 246, groups: id, 82## AIC = 653.4, DIC = 597.1## deviance = 613.3New data
01:00
Load in the three-lev.csv data
threelev <- read_csv(here::here("data", "three-lev.csv"))threelev## # A tibble: 7,230 x 12## schid sid size lowinc mobility female black hispanic retained grade year math## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 2020 273026452 380 40.3 12.5 0 0 1 0 2 0.5 1.146## 2 2020 273026452 380 40.3 12.5 0 0 1 0 3 1.5 1.134## 3 2020 273026452 380 40.3 12.5 0 0 1 0 4 2.5 2.3 ## 4 2020 273030991 380 40.3 12.5 0 0 0 0 0 -1.5 -1.303## 5 2020 273030991 380 40.3 12.5 0 0 0 0 1 -0.5 0.439## 6 2020 273030991 380 40.3 12.5 0 0 0 0 2 0.5 2.43 ## 7 2020 273030991 380 40.3 12.5 0 0 0 0 3 1.5 2.254## 8 2020 273030991 380 40.3 12.5 0 0 0 0 4 2.5 3.873## 9 2020 273059461 380 40.3 12.5 0 0 1 0 0 -1.5 -1.384## 10 2020 273059461 380 40.3 12.5 0 0 1 0 1 -0.5 0.338## # … with 7,220 more rowsRQ 1
To what extent do math scores, and changes in math scores, vary between students versus between schools?
01:00
Two part analysis - first estimate the model, then compute the ICC for the intercept and the ICC for the slope
The model
rq1_math <- lmer(math ~ 1 + year + (year | sid) + (year | schid), data = threelev)Variances
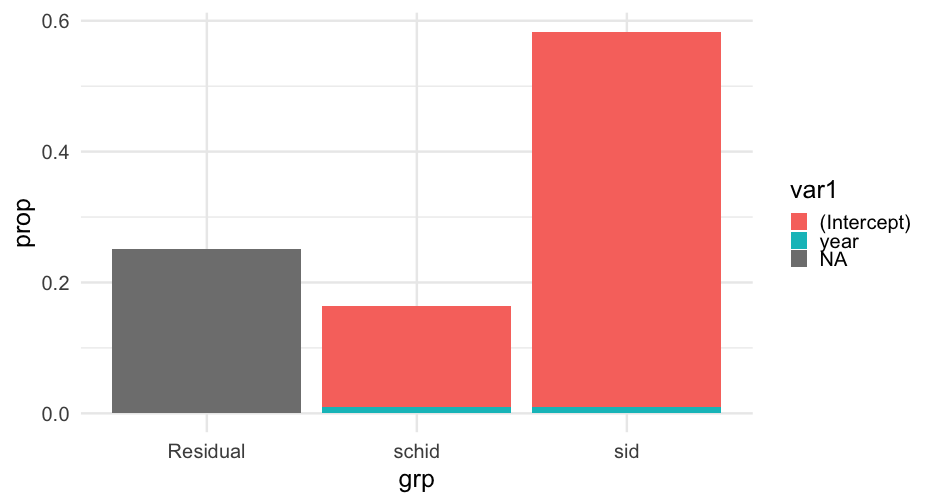
as.data.frame(VarCorr(rq1_math))## grp var1 var2 vcov sdcor## 1 sid (Intercept) <NA> 0.64047114 0.8002944## 2 sid year <NA> 0.01125765 0.1061021## 3 sid (Intercept) year 0.04678624 0.5509911## 4 schid (Intercept) <NA> 0.16857056 0.4105735## 5 schid year <NA> 0.01126367 0.1061304## 6 schid (Intercept) year 0.01734150 0.3979749## 7 Residual <NA> <NA> 0.30143334 0.5490295Proportions
as.data.frame(VarCorr(rq1_math)) %>% mutate(prop = vcov / sum(vcov))## grp var1 var2 vcov sdcor prop## 1 sid (Intercept) <NA> 0.64047114 0.8002944 0.535008142## 2 sid year <NA> 0.01125765 0.1061021 0.009403910## 3 sid (Intercept) year 0.04678624 0.5509911 0.039082201## 4 schid (Intercept) <NA> 0.16857056 0.4105735 0.140812939## 5 schid year <NA> 0.01126367 0.1061304 0.009408942## 6 schid (Intercept) year 0.01734150 0.3979749 0.014485963## 7 Residual <NA> <NA> 0.30143334 0.5490295 0.251797903Last one
02:00
To what extent do the differences in gains between students coded Black or Hispanic, versus those who are not, vary between schools?
Note - this is a model that is difficult to fit. Bayes might help.
rq3_math <- lmer(math ~ year + black + hispanic + year:black + year:hispanic + (year | sid) + (year + year:black + year:hispanic | schid), data = threelev)Low variance components make convergence difficult
arm::display(rq3_math)## lmer(formula = math ~ year + black + hispanic + year:black + ## year:hispanic + (year | sid) + (year + year:black + year:hispanic | ## schid), data = threelev)## coef.est coef.se## (Intercept) -0.36 0.08 ## year 0.78 0.02 ## black -0.59 0.08 ## hispanic -0.38 0.09 ## year:black -0.05 0.02 ## year:hispanic 0.04 0.03 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 0.79 ## year 0.10 0.56 ## schid (Intercept) 0.34 ## year 0.11 0.39 ## year:black 0.07 -0.40 -0.46 ## year:hispanic 0.06 0.03 -0.38 0.90 ## Residual 0.55 ## ---## number of obs: 7230, groups: sid, 1721; schid, 60## AIC = 16327.4, DIC = 16229.3## deviance = 16258.4Reminder
Lung cancer data: Patients nested in doctors
hdp <- read_csv("https://stats.idre.ucla.edu/stat/data/hdp.csv") %>% janitor::clean_names() %>% select(did, tumorsize, pain, lungcapacity, age, remission)hdp## # A tibble: 8,525 x 6## did tumorsize pain lungcapacity age remission## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 67.98120 4 0.8010882 64.96824 0## 2 1 64.70246 2 0.3264440 53.91714 0## 3 1 51.56700 6 0.5650309 53.34730 0## 4 1 86.43799 3 0.8484109 41.36804 0## 5 1 53.40018 3 0.8864910 46.80042 0## 6 1 51.65727 4 0.7010307 51.92936 0## 7 1 78.91707 3 0.8908539 53.82926 0## 8 1 69.83325 3 0.6608795 46.56223 0## 9 1 62.85259 4 0.9088714 54.38936 0## 10 1 71.77790 5 0.9593268 50.54465 0## # … with 8,515 more rowsPredict remission
Build a model where age, lung capacity, and tumor size predict whether or not the patient was in remission.
Build the model so you can evaluate whether or not the relation between the tumor size and likelihood of remission depends on age
Allow the intercept to vary by the doctor ID.
Fit the model using brms
Lung cancer remission model
library(brms)lc <- brm( remission ~ age * tumorsize + lungcapacity + (1|did), data = hdp, family = bernoulli(link = "logit"), cores = 4, backend = "cmdstan")## -\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/Running MCMC with 4 parallel chains...## ## Chain 1 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 2 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 3 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 4 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 3 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 1 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 2 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 4 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 3 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 2 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 1 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 4 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 3 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 1 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 2 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 4 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 3 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 1 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 2 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 4 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 3 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 1 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 2 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 4 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 3 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 1 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 2 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 4 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 3 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 1 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 2 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 4 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 3 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 1 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 2 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 4 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 3 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 1 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 2 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 4 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 3 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 3 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 1 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 1 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 2 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 2 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 4 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 4 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 3 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 1 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 2 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 4 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 3 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 1 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 2 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 4 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 3 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 1 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 2 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 4 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 3 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 1 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 2 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 4 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 3 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 1 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 2 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 4 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 3 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 1 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 2 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 4 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 3 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 1 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 2 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 4 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 3 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 1 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 2 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 4 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 3 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 1 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 2 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 4 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 3 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 3 finished in 307.4 seconds.## Chain 1 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 1 finished in 308.9 seconds.## Chain 2 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 2 finished in 310.6 seconds.## Chain 4 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 4 finished in 315.4 seconds.## ## All 4 chains finished successfully.## Mean chain execution time: 310.6 seconds.## Total execution time: 315.8 seconds.Variance by Doctor
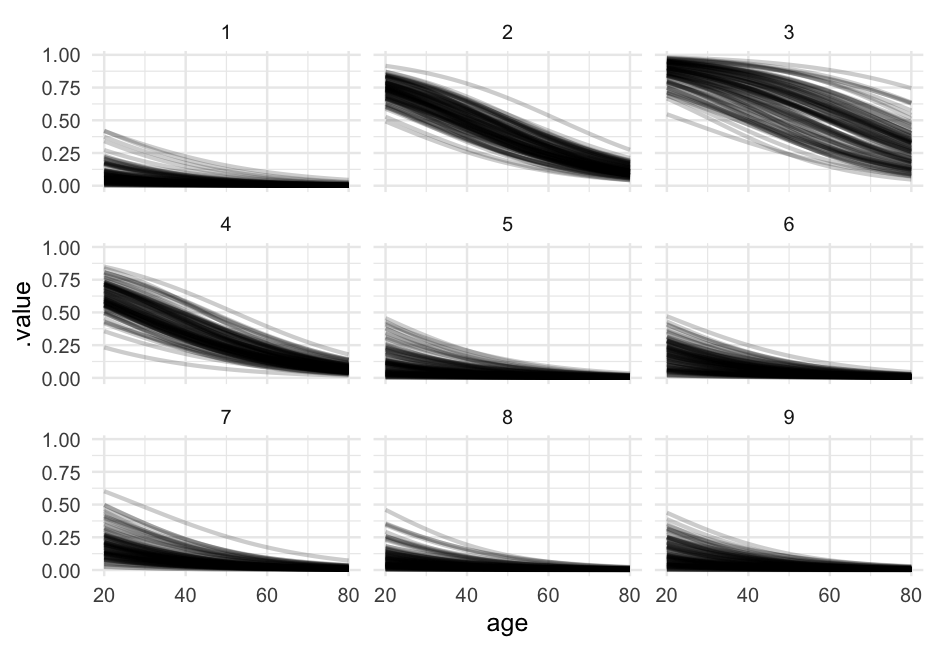
Let's look at the relation between age and probability of remission for each of the first nine doctors.
library(tidybayes)pred_age_doctor <- expand.grid( did = 1:9, age = 20:80, tumorsize = mean(hdp$tumorsize), lungcapacity = mean(hdp$lungcapacity) ) %>% add_fitted_draws(model = lc, n = 100)pred_age_doctor## # A tibble: 54,900 x 9## # Groups: did, age, tumorsize, lungcapacity, .row [549]## did age tumorsize lungcapacity .row .chain .iteration .draw .value## <int> <int> <dbl> <dbl> <int> <int> <int> <int> <dbl>## 1 1 20 70.88067 0.7740865 1 NA NA 7 0.06478647## 2 1 20 70.88067 0.7740865 1 NA NA 13 0.1094038 ## 3 1 20 70.88067 0.7740865 1 NA NA 99 0.07673222## 4 1 20 70.88067 0.7740865 1 NA NA 113 0.06245461## 5 1 20 70.88067 0.7740865 1 NA NA 121 0.02183182## 6 1 20 70.88067 0.7740865 1 NA NA 122 0.1088385 ## 7 1 20 70.88067 0.7740865 1 NA NA 141 0.08642317## 8 1 20 70.88067 0.7740865 1 NA NA 183 0.04355602## 9 1 20 70.88067 0.7740865 1 NA NA 224 0.2211663 ## 10 1 20 70.88067 0.7740865 1 NA NA 288 0.08792308## # … with 54,890 more rowsLook at our variables
get_variables(lc)## [1] "b_Intercept" "b_age" "b_tumorsize" "b_lungcapacity" "b_age:tumorsize" "sd_did__Intercept" ## [7] "Intercept" "r_did[1,Intercept]" "r_did[2,Intercept]" "r_did[3,Intercept]" "r_did[4,Intercept]" "r_did[5,Intercept]" ## [13] "r_did[6,Intercept]" "r_did[7,Intercept]" "r_did[8,Intercept]" "r_did[9,Intercept]" "r_did[10,Intercept]" "r_did[11,Intercept]" ## [19] "r_did[12,Intercept]" "r_did[13,Intercept]" "r_did[14,Intercept]" "r_did[15,Intercept]" "r_did[16,Intercept]" "r_did[17,Intercept]" ## [25] "r_did[18,Intercept]" "r_did[19,Intercept]" "r_did[20,Intercept]" "r_did[21,Intercept]" "r_did[22,Intercept]" "r_did[23,Intercept]" ## [31] "r_did[24,Intercept]" "r_did[25,Intercept]" "r_did[26,Intercept]" "r_did[27,Intercept]" "r_did[28,Intercept]" "r_did[29,Intercept]" ## [37] "r_did[30,Intercept]" "r_did[31,Intercept]" "r_did[32,Intercept]" "r_did[33,Intercept]" "r_did[34,Intercept]" "r_did[35,Intercept]" ## [43] "r_did[36,Intercept]" "r_did[37,Intercept]" "r_did[38,Intercept]" "r_did[39,Intercept]" "r_did[40,Intercept]" "r_did[41,Intercept]" ## [49] "r_did[42,Intercept]" "r_did[43,Intercept]" "r_did[44,Intercept]" "r_did[45,Intercept]" "r_did[46,Intercept]" "r_did[47,Intercept]" ## [55] "r_did[48,Intercept]" "r_did[49,Intercept]" "r_did[50,Intercept]" "r_did[51,Intercept]" "r_did[52,Intercept]" "r_did[53,Intercept]" ## [61] "r_did[54,Intercept]" "r_did[55,Intercept]" "r_did[56,Intercept]" "r_did[57,Intercept]" "r_did[58,Intercept]" "r_did[59,Intercept]" ## [67] "r_did[60,Intercept]" "r_did[61,Intercept]" "r_did[62,Intercept]" "r_did[63,Intercept]" "r_did[64,Intercept]" "r_did[65,Intercept]" ## [73] "r_did[66,Intercept]" "r_did[67,Intercept]" "r_did[68,Intercept]" "r_did[69,Intercept]" "r_did[70,Intercept]" "r_did[71,Intercept]" ## [79] "r_did[72,Intercept]" "r_did[73,Intercept]" "r_did[74,Intercept]" "r_did[75,Intercept]" "r_did[76,Intercept]" "r_did[77,Intercept]" ## [85] "r_did[78,Intercept]" "r_did[79,Intercept]" "r_did[80,Intercept]" "r_did[81,Intercept]" "r_did[82,Intercept]" "r_did[83,Intercept]" ## [91] "r_did[84,Intercept]" "r_did[85,Intercept]" "r_did[86,Intercept]" "r_did[87,Intercept]" "r_did[88,Intercept]" "r_did[89,Intercept]" ## [97] "r_did[90,Intercept]" "r_did[91,Intercept]" "r_did[92,Intercept]" "r_did[93,Intercept]" "r_did[94,Intercept]" "r_did[95,Intercept]" ## [103] "r_did[96,Intercept]" "r_did[97,Intercept]" "r_did[98,Intercept]" "r_did[99,Intercept]" "r_did[100,Intercept]" "r_did[101,Intercept]"## [109] "r_did[102,Intercept]" "r_did[103,Intercept]" "r_did[104,Intercept]" "r_did[105,Intercept]" "r_did[106,Intercept]" "r_did[107,Intercept]"## [115] "r_did[108,Intercept]" "r_did[109,Intercept]" "r_did[110,Intercept]" "r_did[111,Intercept]" "r_did[112,Intercept]" "r_did[113,Intercept]"## [121] "r_did[114,Intercept]" "r_did[115,Intercept]" "r_did[116,Intercept]" "r_did[117,Intercept]" "r_did[118,Intercept]" "r_did[119,Intercept]"## [127] "r_did[120,Intercept]" "r_did[121,Intercept]" "r_did[122,Intercept]" "r_did[123,Intercept]" "r_did[124,Intercept]" "r_did[125,Intercept]"## [133] "r_did[126,Intercept]" "r_did[127,Intercept]" "r_did[128,Intercept]" "r_did[129,Intercept]" "r_did[130,Intercept]" "r_did[131,Intercept]"## [139] "r_did[132,Intercept]" "r_did[133,Intercept]" "r_did[134,Intercept]" "r_did[135,Intercept]" "r_did[136,Intercept]" "r_did[137,Intercept]"## [145] "r_did[138,Intercept]" "r_did[139,Intercept]" "r_did[140,Intercept]" "r_did[141,Intercept]" "r_did[142,Intercept]" "r_did[143,Intercept]"## [151] "r_did[144,Intercept]" "r_did[145,Intercept]" "r_did[146,Intercept]" "r_did[147,Intercept]" "r_did[148,Intercept]" "r_did[149,Intercept]"## [157] "r_did[150,Intercept]" "r_did[151,Intercept]" "r_did[152,Intercept]" "r_did[153,Intercept]" "r_did[154,Intercept]" "r_did[155,Intercept]"## [163] "r_did[156,Intercept]" "r_did[157,Intercept]" "r_did[158,Intercept]" "r_did[159,Intercept]" "r_did[160,Intercept]" "r_did[161,Intercept]"## [169] "r_did[162,Intercept]" "r_did[163,Intercept]" "r_did[164,Intercept]" "r_did[165,Intercept]" "r_did[166,Intercept]" "r_did[167,Intercept]"## [175] "r_did[168,Intercept]" "r_did[169,Intercept]" "r_did[170,Intercept]" "r_did[171,Intercept]" "r_did[172,Intercept]" "r_did[173,Intercept]"## [181] "r_did[174,Intercept]" "r_did[175,Intercept]" "r_did[176,Intercept]" "r_did[177,Intercept]" "r_did[178,Intercept]" "r_did[179,Intercept]"## [187] "r_did[180,Intercept]" "r_did[181,Intercept]" "r_did[182,Intercept]" "r_did[183,Intercept]" "r_did[184,Intercept]" "r_did[185,Intercept]"## [193] "r_did[186,Intercept]" "r_did[187,Intercept]" "r_did[188,Intercept]" "r_did[189,Intercept]" "r_did[190,Intercept]" "r_did[191,Intercept]"## [199] "r_did[192,Intercept]" "r_did[193,Intercept]" "r_did[194,Intercept]" "r_did[195,Intercept]" "r_did[196,Intercept]" "r_did[197,Intercept]"## [205] "r_did[198,Intercept]" "r_did[199,Intercept]" "r_did[200,Intercept]" "r_did[201,Intercept]" "r_did[202,Intercept]" "r_did[203,Intercept]"## [211] "r_did[204,Intercept]" "r_did[205,Intercept]" "r_did[206,Intercept]" "r_did[207,Intercept]" "r_did[208,Intercept]" "r_did[209,Intercept]"## [217] "r_did[210,Intercept]" "r_did[211,Intercept]" "r_did[212,Intercept]" "r_did[213,Intercept]" "r_did[214,Intercept]" "r_did[215,Intercept]"## [223] "r_did[216,Intercept]" "r_did[217,Intercept]" "r_did[218,Intercept]" "r_did[219,Intercept]" "r_did[220,Intercept]" "r_did[221,Intercept]"## [229] "r_did[222,Intercept]" "r_did[223,Intercept]" "r_did[224,Intercept]" "r_did[225,Intercept]" "r_did[226,Intercept]" "r_did[227,Intercept]"## [235] "r_did[228,Intercept]" "r_did[229,Intercept]" "r_did[230,Intercept]" "r_did[231,Intercept]" "r_did[232,Intercept]" "r_did[233,Intercept]"## [241] "r_did[234,Intercept]" "r_did[235,Intercept]" "r_did[236,Intercept]" "r_did[237,Intercept]" "r_did[238,Intercept]" "r_did[239,Intercept]"## [247] "r_did[240,Intercept]" "r_did[241,Intercept]" "r_did[242,Intercept]" "r_did[243,Intercept]" "r_did[244,Intercept]" "r_did[245,Intercept]"## [253] "r_did[246,Intercept]" "r_did[247,Intercept]" "r_did[248,Intercept]" "r_did[249,Intercept]" "r_did[250,Intercept]" "r_did[251,Intercept]"## [259] "r_did[252,Intercept]" "r_did[253,Intercept]" "r_did[254,Intercept]" "r_did[255,Intercept]" "r_did[256,Intercept]" "r_did[257,Intercept]"## [265] "r_did[258,Intercept]" "r_did[259,Intercept]" "r_did[260,Intercept]" "r_did[261,Intercept]" "r_did[262,Intercept]" "r_did[263,Intercept]"## [271] "r_did[264,Intercept]" "r_did[265,Intercept]" "r_did[266,Intercept]" "r_did[267,Intercept]" "r_did[268,Intercept]" "r_did[269,Intercept]"## [277] "r_did[270,Intercept]" "r_did[271,Intercept]" "r_did[272,Intercept]" "r_did[273,Intercept]" "r_did[274,Intercept]" "r_did[275,Intercept]"## [283] "r_did[276,Intercept]" "r_did[277,Intercept]" "r_did[278,Intercept]" "r_did[279,Intercept]" "r_did[280,Intercept]" "r_did[281,Intercept]"## [289] "r_did[282,Intercept]" "r_did[283,Intercept]" "r_did[284,Intercept]" "r_did[285,Intercept]" "r_did[286,Intercept]" "r_did[287,Intercept]"## [295] "r_did[288,Intercept]" "r_did[289,Intercept]" "r_did[290,Intercept]" "r_did[291,Intercept]" "r_did[292,Intercept]" "r_did[293,Intercept]"## [301] "r_did[294,Intercept]" "r_did[295,Intercept]" "r_did[296,Intercept]" "r_did[297,Intercept]" "r_did[298,Intercept]" "r_did[299,Intercept]"## [307] "r_did[300,Intercept]" "r_did[301,Intercept]" "r_did[302,Intercept]" "r_did[303,Intercept]" "r_did[304,Intercept]" "r_did[305,Intercept]"## [313] "r_did[306,Intercept]" "r_did[307,Intercept]" "r_did[308,Intercept]" "r_did[309,Intercept]" "r_did[310,Intercept]" "r_did[311,Intercept]"## [319] "r_did[312,Intercept]" "r_did[313,Intercept]" "r_did[314,Intercept]" "r_did[315,Intercept]" "r_did[316,Intercept]" "r_did[317,Intercept]"## [325] "r_did[318,Intercept]" "r_did[319,Intercept]" "r_did[320,Intercept]" "r_did[321,Intercept]" "r_did[322,Intercept]" "r_did[323,Intercept]"## [331] "r_did[324,Intercept]" "r_did[325,Intercept]" "r_did[326,Intercept]" "r_did[327,Intercept]" "r_did[328,Intercept]" "r_did[329,Intercept]"## [337] "r_did[330,Intercept]" "r_did[331,Intercept]" "r_did[332,Intercept]" "r_did[333,Intercept]" "r_did[334,Intercept]" "r_did[335,Intercept]"## [343] "r_did[336,Intercept]" "r_did[337,Intercept]" "r_did[338,Intercept]" "r_did[339,Intercept]" "r_did[340,Intercept]" "r_did[341,Intercept]"## [349] "r_did[342,Intercept]" "r_did[343,Intercept]" "r_did[344,Intercept]" "r_did[345,Intercept]" "r_did[346,Intercept]" "r_did[347,Intercept]"## [355] "r_did[348,Intercept]" "r_did[349,Intercept]" "r_did[350,Intercept]" "r_did[351,Intercept]" "r_did[352,Intercept]" "r_did[353,Intercept]"## [361] "r_did[354,Intercept]" "r_did[355,Intercept]" "r_did[356,Intercept]" "r_did[357,Intercept]" "r_did[358,Intercept]" "r_did[359,Intercept]"## [367] "r_did[360,Intercept]" "r_did[361,Intercept]" "r_did[362,Intercept]" "r_did[363,Intercept]" "r_did[364,Intercept]" "r_did[365,Intercept]"## [373] "r_did[366,Intercept]" "r_did[367,Intercept]" "r_did[368,Intercept]" "r_did[369,Intercept]" "r_did[370,Intercept]" "r_did[371,Intercept]"## [379] "r_did[372,Intercept]" "r_did[373,Intercept]" "r_did[374,Intercept]" "r_did[375,Intercept]" "r_did[376,Intercept]" "r_did[377,Intercept]"## [385] "r_did[378,Intercept]" "r_did[379,Intercept]" "r_did[380,Intercept]" "r_did[381,Intercept]" "r_did[382,Intercept]" "r_did[383,Intercept]"## [391] "r_did[384,Intercept]" "r_did[385,Intercept]" "r_did[386,Intercept]" "r_did[387,Intercept]" "r_did[388,Intercept]" "r_did[389,Intercept]"## [397] "r_did[390,Intercept]" "r_did[391,Intercept]" "r_did[392,Intercept]" "r_did[393,Intercept]" "r_did[394,Intercept]" "r_did[395,Intercept]"## [403] "r_did[396,Intercept]" "r_did[397,Intercept]" "r_did[398,Intercept]" "r_did[399,Intercept]" "r_did[400,Intercept]" "r_did[401,Intercept]"## [409] "r_did[402,Intercept]" "r_did[403,Intercept]" "r_did[404,Intercept]" "r_did[405,Intercept]" "r_did[406,Intercept]" "r_did[407,Intercept]"## [415] "lp__" "z_1[1,1]" "z_1[1,2]" "z_1[1,3]" "z_1[1,4]" "z_1[1,5]" ## [421] "z_1[1,6]" "z_1[1,7]" "z_1[1,8]" "z_1[1,9]" "z_1[1,10]" "z_1[1,11]" ## [427] "z_1[1,12]" "z_1[1,13]" "z_1[1,14]" "z_1[1,15]" "z_1[1,16]" "z_1[1,17]" ## [433] "z_1[1,18]" "z_1[1,19]" "z_1[1,20]" "z_1[1,21]" "z_1[1,22]" "z_1[1,23]" ## [439] "z_1[1,24]" "z_1[1,25]" "z_1[1,26]" "z_1[1,27]" "z_1[1,28]" "z_1[1,29]" ## [445] "z_1[1,30]" "z_1[1,31]" "z_1[1,32]" "z_1[1,33]" "z_1[1,34]" "z_1[1,35]" ## [451] "z_1[1,36]" "z_1[1,37]" "z_1[1,38]" "z_1[1,39]" "z_1[1,40]" "z_1[1,41]" ## [457] "z_1[1,42]" "z_1[1,43]" "z_1[1,44]" "z_1[1,45]" "z_1[1,46]" "z_1[1,47]" ## [463] "z_1[1,48]" "z_1[1,49]" "z_1[1,50]" "z_1[1,51]" "z_1[1,52]" "z_1[1,53]" ## [469] "z_1[1,54]" "z_1[1,55]" "z_1[1,56]" "z_1[1,57]" "z_1[1,58]" "z_1[1,59]" ## [475] "z_1[1,60]" "z_1[1,61]" "z_1[1,62]" "z_1[1,63]" "z_1[1,64]" "z_1[1,65]" ## [481] "z_1[1,66]" "z_1[1,67]" "z_1[1,68]" "z_1[1,69]" "z_1[1,70]" "z_1[1,71]" ## [487] "z_1[1,72]" "z_1[1,73]" "z_1[1,74]" "z_1[1,75]" "z_1[1,76]" "z_1[1,77]" ## [493] "z_1[1,78]" "z_1[1,79]" "z_1[1,80]" "z_1[1,81]" "z_1[1,82]" "z_1[1,83]" ## [499] "z_1[1,84]" "z_1[1,85]" "z_1[1,86]" "z_1[1,87]" "z_1[1,88]" "z_1[1,89]" ## [505] "z_1[1,90]" "z_1[1,91]" "z_1[1,92]" "z_1[1,93]" "z_1[1,94]" "z_1[1,95]" ## [511] "z_1[1,96]" "z_1[1,97]" "z_1[1,98]" "z_1[1,99]" "z_1[1,100]" "z_1[1,101]" ## [517] "z_1[1,102]" "z_1[1,103]" "z_1[1,104]" "z_1[1,105]" "z_1[1,106]" "z_1[1,107]" ## [523] "z_1[1,108]" "z_1[1,109]" "z_1[1,110]" "z_1[1,111]" "z_1[1,112]" "z_1[1,113]" ## [529] "z_1[1,114]" "z_1[1,115]" "z_1[1,116]" "z_1[1,117]" "z_1[1,118]" "z_1[1,119]" ## [535] "z_1[1,120]" "z_1[1,121]" "z_1[1,122]" "z_1[1,123]" "z_1[1,124]" "z_1[1,125]" ## [541] "z_1[1,126]" "z_1[1,127]" "z_1[1,128]" "z_1[1,129]" "z_1[1,130]" "z_1[1,131]" ## [547] "z_1[1,132]" "z_1[1,133]" "z_1[1,134]" "z_1[1,135]" "z_1[1,136]" "z_1[1,137]" ## [553] "z_1[1,138]" "z_1[1,139]" "z_1[1,140]" "z_1[1,141]" "z_1[1,142]" "z_1[1,143]" ## [559] "z_1[1,144]" "z_1[1,145]" "z_1[1,146]" "z_1[1,147]" "z_1[1,148]" "z_1[1,149]" ## [565] "z_1[1,150]" "z_1[1,151]" "z_1[1,152]" "z_1[1,153]" "z_1[1,154]" "z_1[1,155]" ## [571] "z_1[1,156]" "z_1[1,157]" "z_1[1,158]" "z_1[1,159]" "z_1[1,160]" "z_1[1,161]" ## [577] "z_1[1,162]" "z_1[1,163]" "z_1[1,164]" "z_1[1,165]" "z_1[1,166]" "z_1[1,167]" ## [583] "z_1[1,168]" "z_1[1,169]" "z_1[1,170]" "z_1[1,171]" "z_1[1,172]" "z_1[1,173]" ## [589] "z_1[1,174]" "z_1[1,175]" "z_1[1,176]" "z_1[1,177]" "z_1[1,178]" "z_1[1,179]" ## [595] "z_1[1,180]" "z_1[1,181]" "z_1[1,182]" "z_1[1,183]" "z_1[1,184]" "z_1[1,185]" ## [601] "z_1[1,186]" "z_1[1,187]" "z_1[1,188]" "z_1[1,189]" "z_1[1,190]" "z_1[1,191]" ## [607] "z_1[1,192]" "z_1[1,193]" "z_1[1,194]" "z_1[1,195]" "z_1[1,196]" "z_1[1,197]" ## [613] "z_1[1,198]" "z_1[1,199]" "z_1[1,200]" "z_1[1,201]" "z_1[1,202]" "z_1[1,203]" ## [619] "z_1[1,204]" "z_1[1,205]" "z_1[1,206]" "z_1[1,207]" "z_1[1,208]" "z_1[1,209]" ## [625] "z_1[1,210]" "z_1[1,211]" "z_1[1,212]" "z_1[1,213]" "z_1[1,214]" "z_1[1,215]" ## [631] "z_1[1,216]" "z_1[1,217]" "z_1[1,218]" "z_1[1,219]" "z_1[1,220]" "z_1[1,221]" ## [637] "z_1[1,222]" "z_1[1,223]" "z_1[1,224]" "z_1[1,225]" "z_1[1,226]" "z_1[1,227]" ## [643] "z_1[1,228]" "z_1[1,229]" "z_1[1,230]" "z_1[1,231]" "z_1[1,232]" "z_1[1,233]" ## [649] "z_1[1,234]" "z_1[1,235]" "z_1[1,236]" "z_1[1,237]" "z_1[1,238]" "z_1[1,239]" ## [655] "z_1[1,240]" "z_1[1,241]" "z_1[1,242]" "z_1[1,243]" "z_1[1,244]" "z_1[1,245]" ## [661] "z_1[1,246]" "z_1[1,247]" "z_1[1,248]" "z_1[1,249]" "z_1[1,250]" "z_1[1,251]" ## [667] "z_1[1,252]" "z_1[1,253]" "z_1[1,254]" "z_1[1,255]" "z_1[1,256]" "z_1[1,257]" ## [673] "z_1[1,258]" "z_1[1,259]" "z_1[1,260]" "z_1[1,261]" "z_1[1,262]" "z_1[1,263]" ## [679] "z_1[1,264]" "z_1[1,265]" "z_1[1,266]" "z_1[1,267]" "z_1[1,268]" "z_1[1,269]" ## [685] "z_1[1,270]" "z_1[1,271]" "z_1[1,272]" "z_1[1,273]" "z_1[1,274]" "z_1[1,275]" ## [691] "z_1[1,276]" "z_1[1,277]" "z_1[1,278]" "z_1[1,279]" "z_1[1,280]" "z_1[1,281]" ## [697] "z_1[1,282]" "z_1[1,283]" "z_1[1,284]" "z_1[1,285]" "z_1[1,286]" "z_1[1,287]" ## [703] "z_1[1,288]" "z_1[1,289]" "z_1[1,290]" "z_1[1,291]" "z_1[1,292]" "z_1[1,293]" ## [709] "z_1[1,294]" "z_1[1,295]" "z_1[1,296]" "z_1[1,297]" "z_1[1,298]" "z_1[1,299]" ## [715] "z_1[1,300]" "z_1[1,301]" "z_1[1,302]" "z_1[1,303]" "z_1[1,304]" "z_1[1,305]" ## [721] "z_1[1,306]" "z_1[1,307]" "z_1[1,308]" "z_1[1,309]" "z_1[1,310]" "z_1[1,311]" ## [727] "z_1[1,312]" "z_1[1,313]" "z_1[1,314]" "z_1[1,315]" "z_1[1,316]" "z_1[1,317]" ## [733] "z_1[1,318]" "z_1[1,319]" "z_1[1,320]" "z_1[1,321]" "z_1[1,322]" "z_1[1,323]" ## [739] "z_1[1,324]" "z_1[1,325]" "z_1[1,326]" "z_1[1,327]" "z_1[1,328]" "z_1[1,329]" ## [745] "z_1[1,330]" "z_1[1,331]" "z_1[1,332]" "z_1[1,333]" "z_1[1,334]" "z_1[1,335]" ## [751] "z_1[1,336]" "z_1[1,337]" "z_1[1,338]" "z_1[1,339]" "z_1[1,340]" "z_1[1,341]" ## [757] "z_1[1,342]" "z_1[1,343]" "z_1[1,344]" "z_1[1,345]" "z_1[1,346]" "z_1[1,347]" ## [763] "z_1[1,348]" "z_1[1,349]" "z_1[1,350]" "z_1[1,351]" "z_1[1,352]" "z_1[1,353]" ## [769] "z_1[1,354]" "z_1[1,355]" "z_1[1,356]" "z_1[1,357]" "z_1[1,358]" "z_1[1,359]" ## [775] "z_1[1,360]" "z_1[1,361]" "z_1[1,362]" "z_1[1,363]" "z_1[1,364]" "z_1[1,365]" ## [781] "z_1[1,366]" "z_1[1,367]" "z_1[1,368]" "z_1[1,369]" "z_1[1,370]" "z_1[1,371]" ## [787] "z_1[1,372]" "z_1[1,373]" "z_1[1,374]" "z_1[1,375]" "z_1[1,376]" "z_1[1,377]" ## [793] "z_1[1,378]" "z_1[1,379]" "z_1[1,380]" "z_1[1,381]" "z_1[1,382]" "z_1[1,383]" ## [799] "z_1[1,384]" "z_1[1,385]" "z_1[1,386]" "z_1[1,387]" "z_1[1,388]" "z_1[1,389]" ## [805] "z_1[1,390]" "z_1[1,391]" "z_1[1,392]" "z_1[1,393]" "z_1[1,394]" "z_1[1,395]" ## [811] "z_1[1,396]" "z_1[1,397]" "z_1[1,398]" "z_1[1,399]" "z_1[1,400]" "z_1[1,401]" ## [817] "z_1[1,402]" "z_1[1,403]" "z_1[1,404]" "z_1[1,405]" "z_1[1,406]" "z_1[1,407]" ## [823] "accept_stat__" "treedepth__" "stepsize__" "divergent__" "n_leapfrog__" "energy__"Multiple comparisons
One of the nicest things about Bayes is that any comparison you want to make can be made without jumping through a lot of additional hoops (e.g., adjusting α).
Scenario
Imagine a 35 year old has a tumor measuring 58 millimeters and a lung capacity rating of 0.81.
What would we estimate as the probability of remission if this patient had did == 1 versus did == 2?
Fixed effects
Not really "fixed", but rather just average relation
fe <- lc %>% spread_draws(b_Intercept, b_age, b_tumorsize, b_lungcapacity, `b_age:tumorsize`)fe## # A tibble: 4,000 x 8## .chain .iteration .draw b_Intercept b_age b_tumorsize b_lungcapacity `b_age:tumorsize`## <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 1 1 5.42858 -0.119206 -0.0544173 -0.362963 0.000883342 ## 2 1 2 2 4.46859 -0.100647 -0.0450108 -0.341147 0.000686135 ## 3 1 3 3 1.96492 -0.0468593 -0.0102471 -0.0702373 -0.000100602 ## 4 1 4 4 0.917227 -0.0301529 0.00952746 -0.0681794 -0.00045534 ## 5 1 5 5 1.53674 -0.0396987 -0.00207063 -0.067519 -0.000231016 ## 6 1 6 6 0.881689 -0.0345708 0.00102342 0.0516893 -0.000215422 ## 7 1 7 7 1.64537 -0.0490206 -0.00906014 0.144082 -0.0000381692## 8 1 8 8 1.38195 -0.0418099 -0.00452064 -0.0829176 -0.000118016 ## 9 1 9 9 0.580196 -0.0293446 0.00953349 -0.214873 -0.000299422 ## 10 1 10 10 3.48012 -0.0843227 -0.0282885 0.226778 0.000286907 ## # … with 3,990 more rowsData
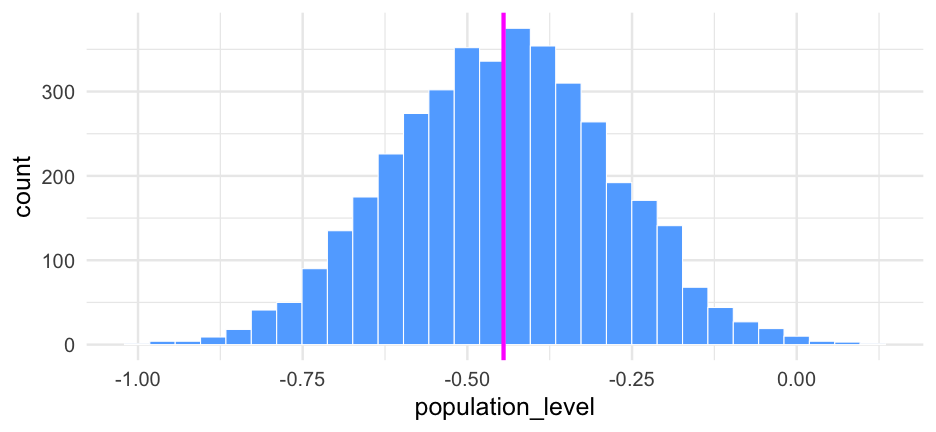
age <- 35tumor_size <- 58lung_cap <- 0.81Population-level predictions (there's other ways we could do this, but it's good to remind ourselves the "by hand" version too)
pop_level <- fe$b_Intercept + (fe$b_age * age) + (fe$b_tumorsize * tumor_size) + (fe$b_lungcapacity * lung_cap) + (fe$`b_age:tumorsize` * (age * tumor_size))pop_level## [1] -0.400649170 -0.548156420 -0.530601573 -0.565097334 -0.496463910 -0.664368967 -0.556616176 -0.650329356 -0.675796370 -0.345796110 -0.220703360## [12] -0.273899070 -0.164508090 -0.505821720 -0.786540482 -0.641475783 -0.729599967 -0.682317970 -0.655076620 -0.688422860 -0.376667380 -0.204903298## [23] -0.276976520 -0.574581568 -0.613817338 -0.457796800 -0.571542413 -0.215873270 -0.390511370 -0.688152020 -0.455349140 -0.315346911 -0.512989300## [34] -0.394965420 -0.272091990 -0.233677040 -0.145021650 -0.203014758 -0.183764710 -0.278536550 -0.181634380 -0.266059696 -0.455146828 -0.371576567## [45] -0.336482186 -0.669924550 -0.404196743 -0.444949890 -0.527445799 -0.393590465 -0.510502935 -0.558348474 -0.611188560 -0.609102980 -0.552765323## [56] -0.687053742 -0.717599319 -0.320627736 -0.566979022 -0.750385008 -0.434146624 -0.605748530 -0.395129009 -0.722624360 -0.366559911 -0.498482117## [67] -0.361466579 -0.454320530 -0.695275430 -0.327491690 -0.699877210 -0.648048410 -0.582501470 -0.727999660 -0.540643810 -0.639119230 -0.537731467## [78] -0.534623838 -0.392257258 -0.636703666 -0.339796006 -0.503746806 -0.636242720 -0.498137280 -0.700199592 -0.657733420 -0.610232197 -0.597943932## [89] -0.726239759 -0.421292220 -0.246458520 -0.337559855 -0.466843460 -0.137523754 -0.422676080 -0.249349500 -0.605182628 -0.231496790 -0.097505142## [100] -0.343801196 -0.263980024 -0.233516836 -0.286142031 -0.173861173 -0.188169444 -0.303025280 -0.032030403 -0.220309850 -0.232198760 -0.569226610## [111] -0.276779560 -0.416181050 -0.513572690 -0.553591230 -0.383143408 -0.455397395 -0.294004800 -0.287911180 -0.750913889 -0.482584865 -0.280293270## [122] -0.419912460 -0.497267460 -0.752613303 -0.762468564 -0.811679146 -0.472403280 -0.629187705 -0.692160511 -0.636007950 -0.556165568 -0.333276159## [133] -0.184457560 -0.110240520 -0.592849956 -0.379310790 -0.632538686 -0.349918793 -0.283377533 -0.200275843 -0.337674390 -0.384618969 -0.530156450## [144] -0.235519500 -0.425610052 -0.216854550 -0.256291660 -0.442811110 -0.274071231 -0.233626365 -0.175077490 -0.420423827 -0.142113530 -0.127813000## [155] -0.192821748 -0.526777810 -0.470034302 -0.418321510 -0.342233425 -0.446490500 -0.672814190 -0.263030610 -0.253975820 -0.494459150 -0.042397740## [166] -0.098761040 -0.266945882 -0.484075070 -0.587978770 -0.489440546 -0.482986944 -0.465884600 -0.096774206 -0.589693990 -0.514907174 -0.546068370## [177] -0.444208850 -0.678808696 -0.582087640 -0.457163094 -0.620135730 -0.548393908 -0.656352616 -0.601464380 -0.618336179 -0.683800078 -0.520241363## [188] -0.510682373 -0.567875110 -0.493480830 -0.556140334 -0.484403090 -0.708244830 -0.361461670 -0.483136344 -0.563771650 -0.293549266 -0.599080620## [199] -0.439578000 -0.476518090 -0.339541300 -0.567153785 -0.577976190 -0.491558986 -0.597674189 -0.528232260 -0.789822866 -0.412859940 -0.506318800## [210] -0.568033920 -0.709689180 -0.629946580 -0.558956683 -0.381128410 -0.345745570 -0.519005497 -0.453566195 -0.427913650 -0.657492740 -0.764623220## [221] -0.403500760 -0.378605098 -0.400736644 -0.359818358 -0.638074178 -0.439748870 -0.468947074 -0.352140100 -0.429509055 -0.538888884 -0.632757935## [232] -0.545479545 -0.544267040 -0.431984980 -0.390400630 -0.481067680 -0.619330659 -0.894500506 -0.334125085 -0.748613205 -0.569310228 -0.434831810## [243] -0.352281370 -0.561997741 -0.290861816 -0.433829791 -0.489476470 -0.635792846 -0.534782281 -0.533142688 -0.559287542 -0.661736550 -0.299543496## [254] -0.257730670 -0.354417643 -0.753819380 -0.389839339 -0.460313492 -0.307565738 -0.622154840 -0.479978040 -0.482922798 -0.772672923 -0.476916610## [265] -0.708744980 -0.559365322 -0.704364700 -0.409778980 -0.624927135 -0.399139126 -0.405146588 -0.506358050 -0.405103074 -0.480418503 -0.589490620## [276] -0.456212740 -0.305062017 -0.404094841 -0.323605880 0.045329230 -0.379168722 -0.526575810 -0.454566440 -0.443256780 -0.641608243 -0.698302880## [287] -0.564627530 -0.423616046 -0.611965186 -0.648255250 -0.394377130 -0.740395869 -0.300188773 -0.327572080 -0.386176050 -0.300323910 -0.355447020## [298] -0.553153200 -0.461874635 -0.342880860 -0.378370097 -0.459946440 -0.417662460 -0.267415990 -0.365536480 -0.530489239 -0.605929932 -0.502403600## [309] -0.642126265 -0.490539665 -0.566497244 -0.437266490 -0.308180250 -0.419428380 -0.155743960 -0.328806570 -0.608592040 -0.236346398 -0.582906559## [320] -0.654138795 -0.613598003 -0.542439290 -0.285669199 -0.675333716 -0.329189559 -0.511355302 -0.557850824 -0.427583600 -0.580947190 -0.491400039## [331] -0.475511683 -0.568146465 -0.344819860 -0.614445404 -0.719698680 -0.293191880 -0.698203350 -0.354556579 -0.428343720 -0.312109350 -0.414847118## [342] -0.168118540 -0.392295237 -0.338308160 -0.284403631 -0.480786950 -0.693700539 -0.327745208 -0.638420878 -0.267827270 -0.205584650 -0.438577840## [353] -0.727853940 -0.561525501 -0.552455120 -0.469068310 -0.474874480 -0.548947370 -0.419293410 -0.322006377 -0.201100620 -0.093800720 -0.238475350## [364] -0.265195160 -0.311731156 -0.435253489 -0.202841100 -0.533999580 -0.417169700 -0.377753480 -0.654115510 -0.104062710 -0.190376930 -0.402292140## [375] -0.418426118 -0.275388830 -0.227262050 -0.280687270 -0.367984910 -0.183319770 -0.007264098 -0.096811290 -0.215860660 -0.130297640 -0.183630140## [386] -0.198312380 -0.577452788 -0.180311920 -0.558406140 -0.400926496 -0.417880485 -0.378693350 -0.445091352 -0.454437524 -0.233878910 -0.225909100## [397] -0.255525712 -0.367465690 -0.385366930 -0.482649302 -0.758117140 -0.504234170 -0.600306940 -0.540933480 -0.498496360 -0.432594898 -0.242141164## [408] -0.413760510 -0.507706626 -0.531098800 -0.391302482 -0.191577260 -0.061849800 -0.194930870 -0.297269215 -0.458989837 -0.171697206 -0.167128180## [419] -0.318776409 -0.512359394 -0.394426660 -0.320670500 -0.555739913 -0.568208094 -0.626846305 -0.681367150 -0.394558490 -0.268698725 -0.232360236## [430] -0.161236837 -0.349186860 -0.265890770 -0.314474100 -0.431511670 -0.227335901 -0.417871610 -0.472181773 -0.335758265 -0.571938906 -0.619151053## [441] -0.557645511 -0.763246258 -0.488016788 -0.451866390 -0.625667710 -0.413880892 -0.570468620 -0.186087240 -0.545170858 -0.632513998 -0.630178080## [452] -0.539907520 -0.725021300 -0.498250612 -0.566659805 -0.670559950 -0.339739290 -0.293468390 -0.664254969 -0.621559928 -0.491867769 -0.764686020## [463] -0.610544774 -0.674888622 -0.257444124 -0.484777340 -0.436602910 -0.440213423 -0.490206860 -0.470335270 -0.714676530 -0.567003900 -0.412001126## [474] -0.424820845 -0.531595960 -0.517873504 -0.468073751 -0.517430213 -0.280402200 -0.486713746 -0.555527650 -0.661969663 -0.775726310 -0.442118250## [485] -0.521893260 -0.437583929 -0.492164871 -0.311701228 -0.345844840 -0.418295633 -0.430895250 -0.527248790 -0.222535600 -0.296124867 -0.261954262## [496] -0.439051422 -0.388819830 -0.497986480 -0.507848277 -0.541291980 -0.777091924 -0.293792634 -0.463545138 -0.250086597 -0.406059172 -0.499645814## [507] -0.396161974 -0.293766973 -0.170887075 -0.597236613 -0.729486800 -0.352659016 -0.050892300 -0.425226280 -0.498033460 -0.111896190 -0.202634945## [518] -0.412666700 -0.498851920 -0.197408290 -0.706957480 -0.760794550 -0.675023080 -0.622399280 -0.594468366 -0.443515745 -0.492007245 -0.410421319## [529] -0.532689839 -0.582932890 -0.780821890 -0.832204589 -0.774849890 -0.674743624 -0.341067895 -0.172593500 -0.101943400 -0.272058129 -0.219886600## [540] -0.158479166 -0.314123793 -0.509055220 -0.411807690 -0.188653753 -0.056500327 -0.071296050 -0.563878317 -0.536439088 -0.555655310 -0.183550780## [551] -0.337437512 -0.481362650 -0.224269384 -0.206933750 -0.241354510 -0.257079213 0.071426914 -0.579425140 -0.336005300 -0.360261710 -0.329678078## [562] -0.573453550 -0.544088070 -0.241598590 -0.353160620 -0.324634400 -0.525225130 -0.698469130 -0.811520100 -0.484320380 -0.406120170 -0.432119410## [573] -0.830096633 -0.478114462 -0.490004960 -0.397325174 -0.612548834 -0.332679200 -0.349292602 -0.470376370 -0.429495500 -0.313588504 -0.295238032## [584] -0.419262990 -0.396212379 -0.401741564 -0.272097940 -0.495632710 -0.540815428 -0.563073324 -0.473583034 -0.321547260 -0.455841369 -0.451252872## [595] -0.408369543 -0.447111110 -0.534860595 -0.550440594 -0.654053272 -0.289423310 -0.456853820 -0.502451770 -0.230975073 -0.420630188 -0.430613400## [606] -0.643020936 -0.307831872 -0.448557720 -0.420799046 -0.377986733 -0.229628640 -0.270418923 -0.208753881 -0.453813781 -0.641687101 -0.401505894## [617] -0.566157590 -0.149189248 -0.213319252 -0.415699689 -0.212954320 -0.798144766 -0.617633800 -0.515462248 -0.332516351 -0.546854260 -0.372998430## [628] -0.698467540 -0.390204220 -0.363271880 -0.289090429 -0.104798600 -0.001310409 -0.455654260 -0.597405142 -0.509213638 -0.481339320 -0.410391000## [639] -0.283276060 -0.482653720 -0.291311820 -0.127802140 -0.253612380 -0.344326353 -0.474444856 -0.254837180 -0.409049087 -0.256853350 -0.236738980## [650] -0.246068667 -0.504758050 -0.441247240 -0.249725710 -0.248071666 -0.334610741 -0.099376380 -0.237671910 -0.192749004 -0.177994220 -0.286501900## [661] -0.376740130 -0.318407210 -0.525148350 -0.437265860 -0.493354832 -0.573574420 -0.553769396 -0.399982630 -0.394562202 -0.421673160 -0.316397880## [672] -0.364705694 -0.338700250 -0.354331297 -0.155571850 -0.711699500 -0.698265185 -0.310253290 -0.290834819 -0.256667690 -0.252584660 -0.658249360## [683] -0.601109980 -0.522455771 -0.727923400 -0.264090770 -0.273555610 -0.333960910 -0.269574490 -0.629687400 -0.474618119 -0.381780800 -0.323293910## [694] -0.377459640 -0.247196237 -0.585836510 -0.576474557 -0.410631840 -0.517714597 -0.243031910 -0.345779922 -0.452710657 -0.273762816 -0.318338320## [705] -0.335733600 -0.577219760 -0.491409558 -0.442086620 -0.438600600 -0.421928506 -0.297823536 -0.327041158 -0.355197794 -0.051137076 -0.353718770## [716] -0.442112643 -0.299483129 -0.560832140 -0.598332860 -0.419798380 -0.372946550 -0.546518210 -0.474653100 -0.424513830 -0.339915820 -0.466266050## [727] -0.418886130 -0.269096750 -0.441293520 -0.382426530 -0.286564399 -0.160431539 -0.562035720 -0.402267880 -0.635643340 -0.495331068 -0.461405210## [738] -0.500835260 -0.344654030 -0.271054910 -0.389202503 -0.490084910 -0.412716070 -0.509191637 -0.483107038 -0.381242960 -0.449640720 -0.218809210## [749] -0.336881710 -0.216061832 -0.204070390 -0.337209970 -0.386444960 -0.197177959 -0.358340781 -0.417889367 -0.599563290 -0.640522200 -0.260670200## [760] -0.217066520 -0.336431620 -0.295897830 -0.550083060 -0.694274429 -0.689812810 -0.447461360 -0.209645600 -0.431500480 -0.503396900 -0.318371990## [771] -0.374558120 -0.377241475 -0.341293069 -0.219982230 -0.358334380 -0.428225700 -0.573135930 -0.548659540 -0.530192620 -0.698215580 -0.486480840## [782] -0.292892145 -0.090415415 -0.464001088 -0.461043470 -0.372778421 -0.433174704 -0.530039650 -0.793809970 -0.668267520 -0.798465220 -0.394810780## [793] -0.616772360 -0.520586062 -0.482716548 -0.188173040 -0.496278131 -0.481497390 -0.591774010 -0.410886397 -0.149240331 -0.111665266 -0.233938620## [804] -0.149136978 -0.313818570 -0.557459630 -0.196040833 -0.306646730 -0.333133085 -0.366021900 -0.311938550 -0.235910490 -0.343918510 -0.322717140## [815] -0.419738282 -0.248630470 -0.564370970 -0.523834824 -0.443765400 -0.410608042 -0.500153567 -0.510730040 -0.351680557 -0.175499822 -0.301668757## [826] -0.270964974 -0.358150567 -0.303199863 -0.268672115 -0.297534728 -0.250162270 -0.465268184 -0.302521240 -0.413297720 -0.899366160 -0.749663260## [837] -0.812177490 -0.605904210 -0.566500500 -0.556802504 -0.526278232 -0.111746319 -0.323026772 -0.355729803 -0.538080910 -0.455716047 -0.259072993## [848] -0.269404033 -0.346585590 -0.263733270 -0.242170560 -0.283107640 -0.406385210 -0.311194350 -0.531103840 -0.617519156 -0.368744260 -0.598699360## [859] -0.539240920 -0.537783657 -0.333762490 -0.412108290 -0.357758246 -0.328603459 -0.240855984 -0.049896140 -0.255521780 -0.417530430 -0.322010346## [870] -0.249250050 -0.246893953 -0.381736249 -0.259369157 -0.439738298 -0.325141170 -0.086054736 -0.156740040 -0.223647770 -0.441732800 -0.352710480## [881] -0.469320791 -0.536869329 -0.502338507 -0.438074480 -0.447929615 -0.528310927 -0.766797779 -0.338437066 -0.201393380 -0.351651460 -0.456395311## [892] -0.608107250 -0.573260190 -0.361135410 -0.558541930 -0.389841945 -0.551656610 -0.632961130 -0.180036365 -0.235444925 -0.482375275 -0.529977320## [903] -0.533778389 -0.616013580 -0.920932230 -0.599693020 -0.652267395 -0.761873290 -0.769476960 -0.657775780 -0.527387626 -0.291910325 -0.328579732## [914] -0.240224257 -0.400291863 -0.298506984 -0.321136510 -0.400663820 -0.180676660 -0.520290927 -0.292328011 -0.525941285 -0.368725900 -0.374598878## [925] -0.223856550 -0.501244710 -0.234560650 -0.338450370 -0.258506740 -0.542715040 -0.387152600 -0.579475540 -0.421962890 -0.411797150 -0.293626301## [936] -0.329830910 -0.288969830 -0.125361620 -0.187621503 -0.550841466 -0.411523570 -0.429196624 -0.474083469 -0.428257296 -0.384163522 -0.460986040## [947] -0.525654280 -0.604416009 -0.497500660 -0.385821815 -0.727306351 -0.557767536 -0.428750051 -0.696497521 -0.563040032 -0.586066234 -0.597437800## [958] -0.437735406 -0.441873670 -0.697801635 -0.834843540 -0.664305660 -0.712805760 -0.624161400 -0.628154010 -0.688781540 -0.455268360 -0.336192508## [969] -0.453272450 -0.433895820 -0.389228650 -0.373178940 -0.401526010 -0.461350538 -0.094635754 -0.276743850 -0.233175625 -0.516766681 -0.336118760## [980] -0.484906016 -0.381328261 -0.282499098 -0.499161220 -0.337716004 -0.483572486 -0.351755708 -0.290827456 -0.316920820 -0.396101823 -0.366105387## [991] -0.284001170 -0.424119550 -0.463104670 -0.773033622 -0.725296690 -0.724969380 -0.366602030 -0.442873242 -0.548111482 -0.629405443## [ reached getOption("max.print") -- omitted 3000 entries ]Transform
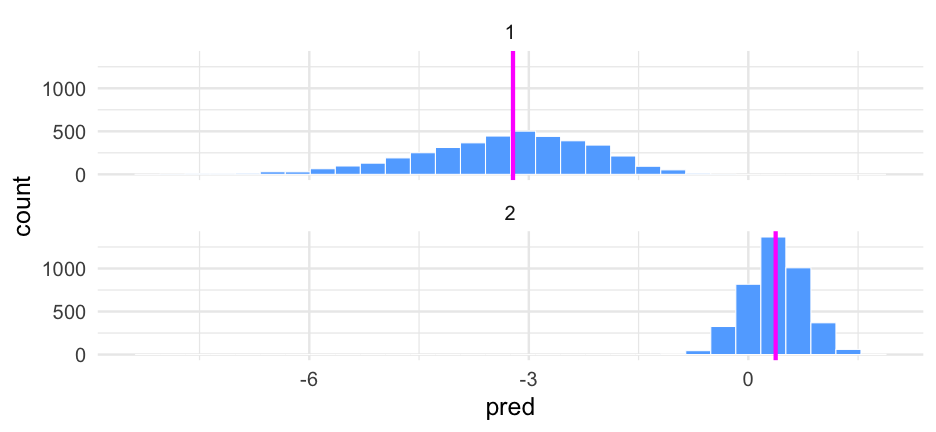
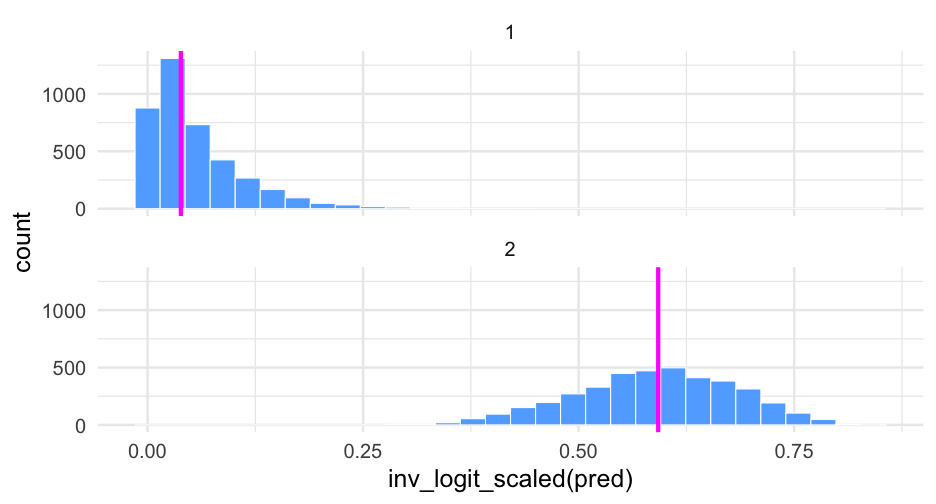
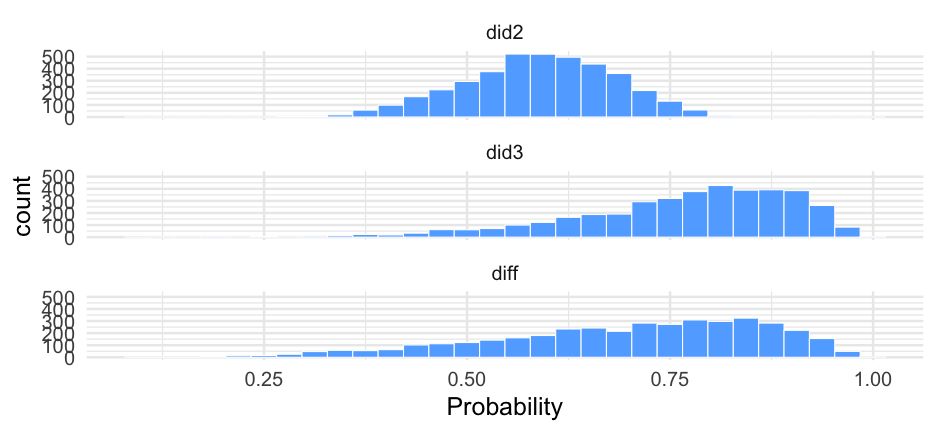
Let's look at this again on the probability scale using brms::inv_logit_scaled() to make the transformation.
ggplot(did12, aes(inv_logit_scaled(pred))) + geom_histogram(fill = "#61adff", color = "white") + geom_vline(aes(xintercept = inv_logit_scaled(did_median)), data = did12_medians, color = "magenta", size = 2) + facet_wrap(~did, ncol = 1)Exponentiation
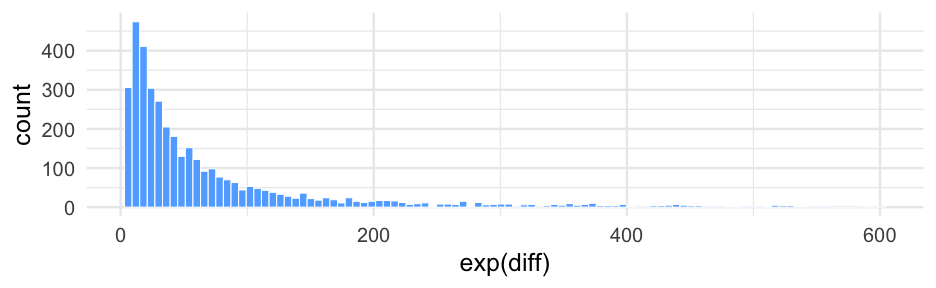
We can exponentiate the log-odds to get normal odds
These are fairly interpretable (especially when greater than 1)
# probabilityinv_logit_scaled(did12_medians$did_median)## [1] 0.03856683 0.59228985# oddsexp(did12_medians$did_median)## [1] 0.0401139 1.4527229# odds of the differenceexp(diff(did12_medians$did_median))## [1] 36.21495Confidence in difference?
Everything is a distribution
Just compute the difference in these distributions, and we get a new distribution, which we can use to summarize our uncertainty
did12_wider <- tibble( did1 = pred_did1, did2 = pred_did2) %>% mutate(diff = did2 - did1)did12_wider## # A tibble: 4,000 x 3## did1 did2 diff## <dbl> <dbl> <dbl>## 1 -4.732689 1.005471 5.73816 ## 2 -4.845796 0.8612836 5.70708 ## 3 -2.986972 0.4373794 3.424351## 4 -0.9940453 0.1152017 1.109247## 5 -5.773234 0.1754881 5.948722## 6 -3.162519 0.6139410 3.77646 ## 7 -3.306486 0.6795638 3.98605 ## 8 -5.793359 0.3006396 6.093999## 9 -4.294496 -0.07275537 4.221741## 10 -1.697236 0.8960439 2.59328 ## # … with 3,990 more rowsDirectionality
Let's say we want to simplify the question to directionality.
Is there a greater chance of remission for did 2 than 1?
table(did12_wider$diff > 0) / 4000## ## TRUE ## 1The distributions are not overlapping at all - therefore, we are as certain as we can be that the odds of remission are higher with did 2 than 1.
One more quick example
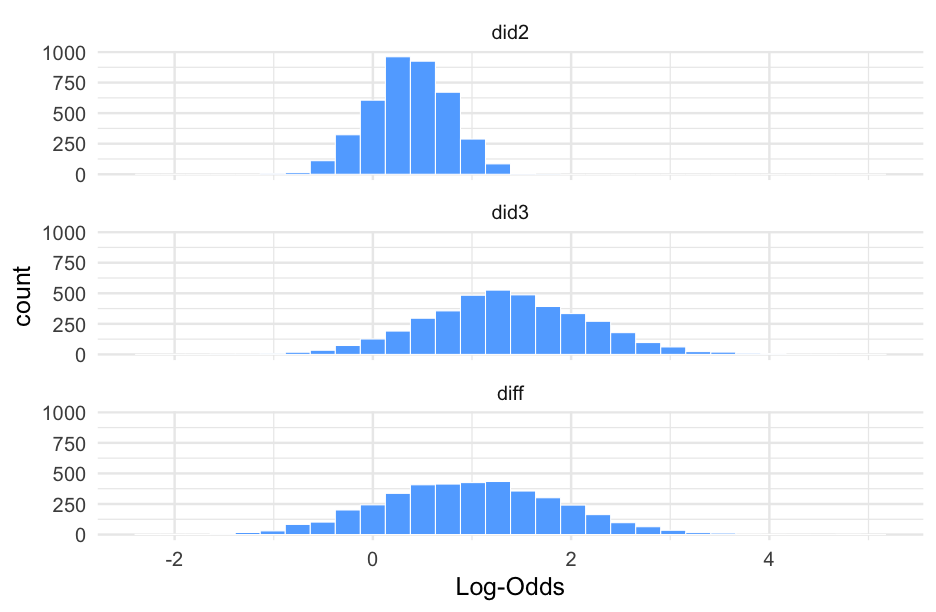
Let's do the same thing, but comparing did 2 and 3.
did3 <- filter(dids, did == 3)pred_did3 <- pop_level + did3$r_diddid23 <- did12_wider %>% select(-did1, -diff) %>% mutate(did3 = pred_did3, diff = did3 - did2)did23## # A tibble: 4,000 x 3## did2 did3 diff## <dbl> <dbl> <dbl>## 1 1.005471 0.8228108 -0.18266 ## 2 0.8612836 0.8277436 -0.03354000## 3 0.4373794 1.004198 0.566819 ## 4 0.1152017 1.194783 1.079581 ## 5 0.1754881 1.432726 1.257238 ## 6 0.6139410 1.354041 0.7401000 ## 7 0.6795638 2.767964 2.0884 ## 8 0.3006396 1.548721 1.248081 ## 9 -0.07275537 2.912974 2.985729 ## 10 0.8960439 -0.06564811 -0.961692 ## # … with 3,990 more rowsPlot data
pd23 <- did23 %>% pivot_longer(did2:diff, names_to = "Distribution", values_to = "Log-Odds")pd23## # A tibble: 12,000 x 2## Distribution `Log-Odds`## <chr> <dbl>## 1 did2 1.005471 ## 2 did3 0.8228108 ## 3 diff -0.18266 ## 4 did2 0.8612836 ## 5 did3 0.8277436 ## 6 diff -0.03354000## 7 did2 0.4373794 ## 8 did3 1.004198 ## 9 diff 0.566819 ## 10 did2 0.1152017 ## # … with 11,990 more rowsMissing data on the IVs
Much more problematic, no matter the model or application
Remove all cases with any missingness on any IV?
Limits your sample size
Might (probably?) introduces new sources of bias
Impute?
- Often ethical challenges here - do you really want to impute somebody's gender?
Solution?
There really isn't a great one. Be clear about the decisions you do make.
If you do choose imputation, use multiple imputation
- This will allow you to have uncertainty in your imputation
The purpose is to get unbiased population estimates for your parameters (not make inferences about an individual for whom data were imputed)
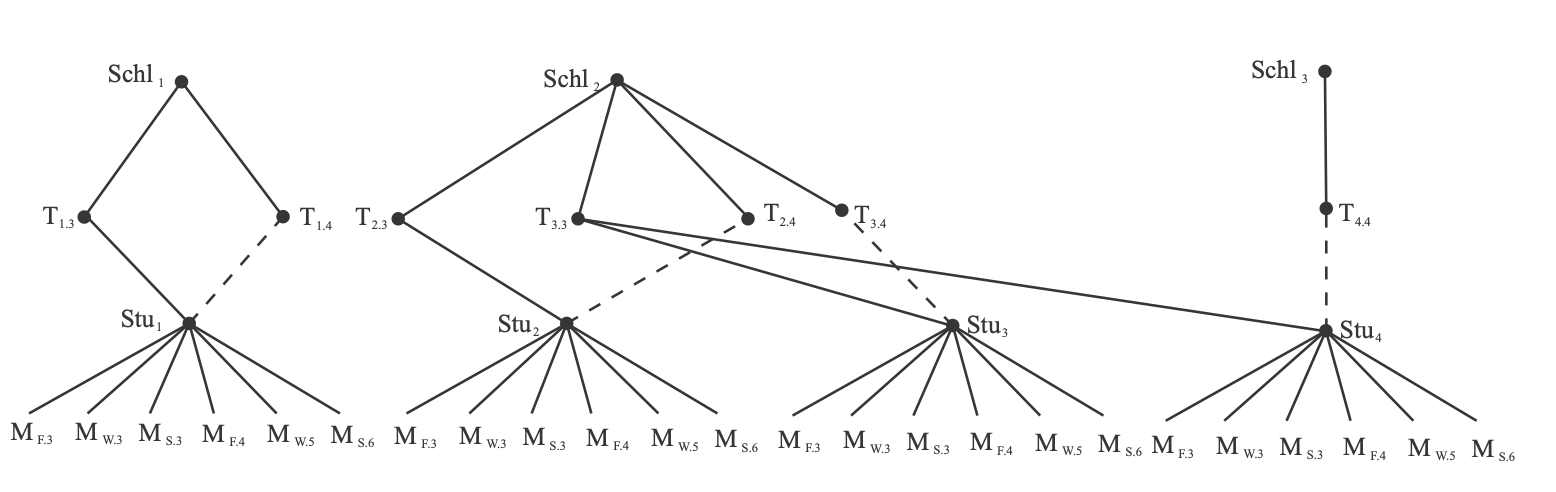
Missing IDs
In multilevel models, you always have IDs linking the data to the higher levels
If you are missing these IDs, I'm not really sure what to tell you
This is particularly common with longitudinal data (e.g., missing prior school IDs)
In rare cases, you can make assumptions and impute, but those are few and far between, in my experience, and the assumptions are still pretty dangerous
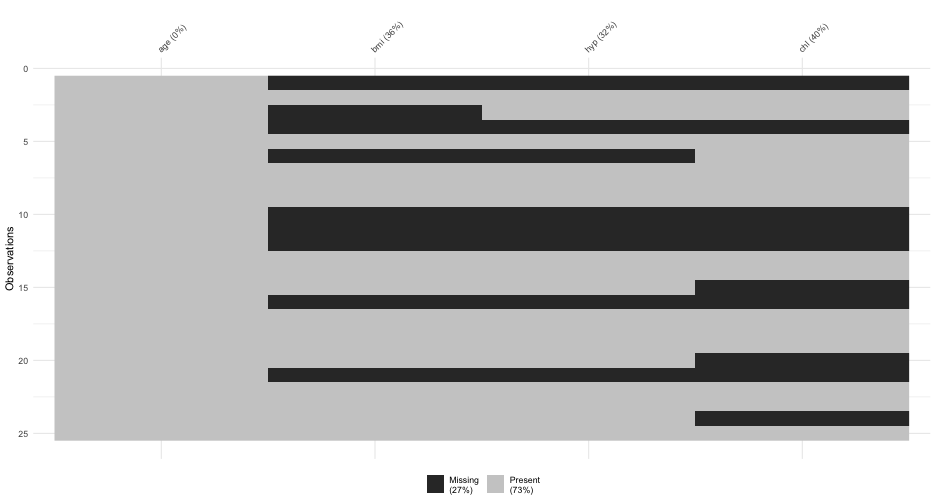
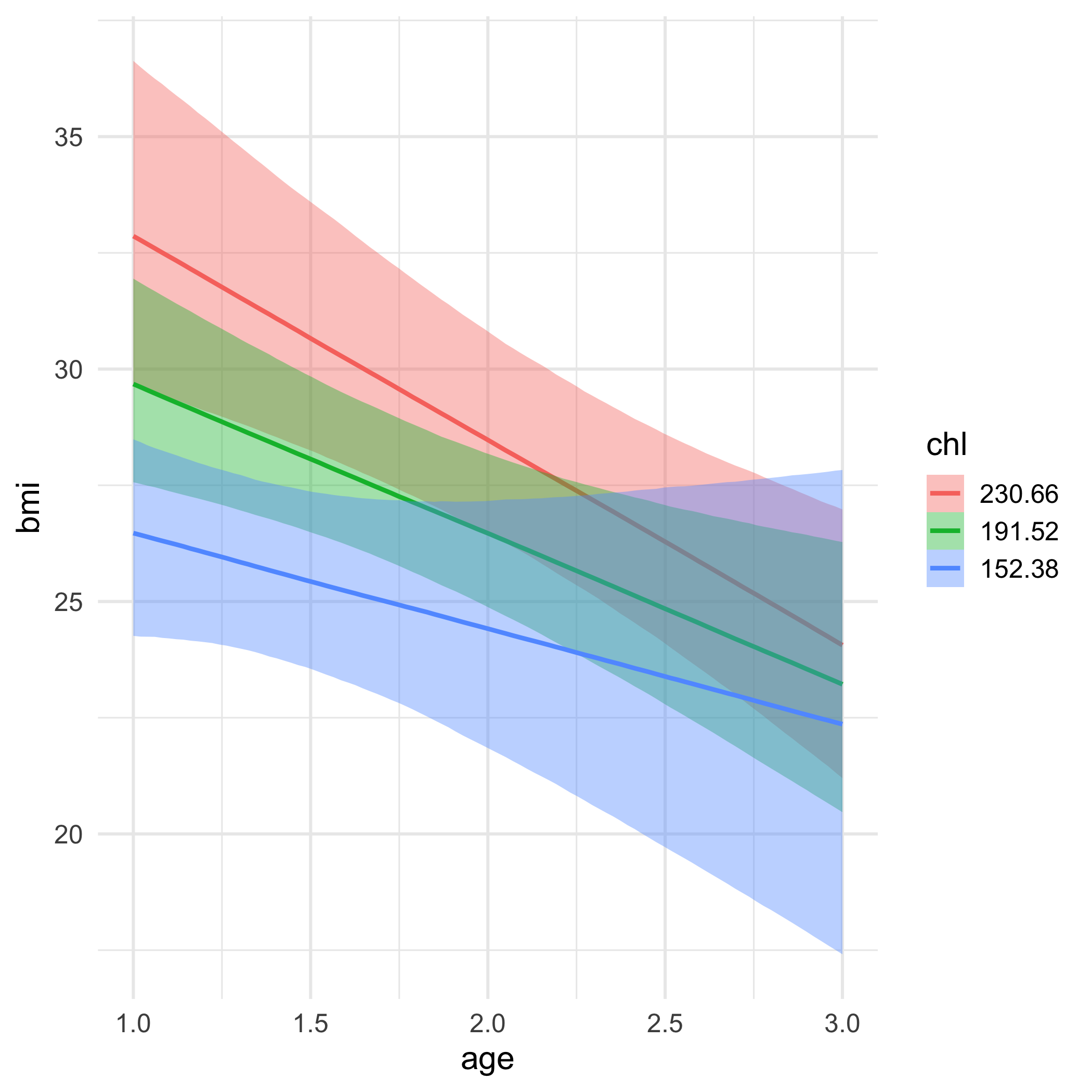
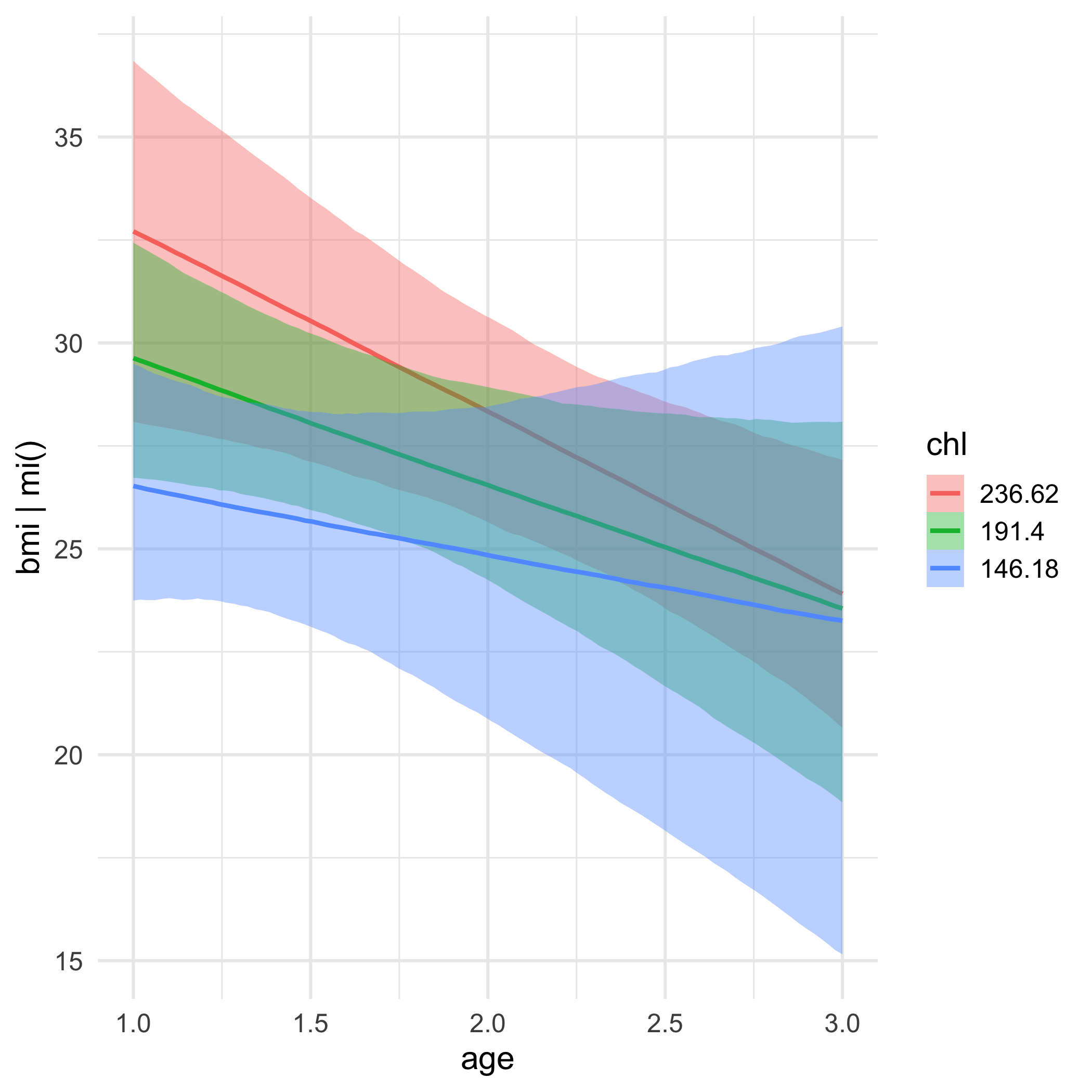
MI for BMI
mi_nhanes$imp$bmi## 1 2 3 4 5## 1 30.1 22.7 35.3 30.1 30.1## 3 27.2 22.0 33.2 29.6 26.3## 4 22.5 24.9 27.4 21.7 21.7## 6 22.5 20.4 22.5 21.7 20.4## 10 27.4 25.5 27.2 20.4 33.2## 11 20.4 27.5 22.5 29.6 22.5## 12 25.5 27.5 24.9 27.5 33.2## 16 27.2 25.5 27.4 35.3 22.0## 21 22.5 35.3 35.3 30.1 30.1Missing formula
We specify a model for each column that has missingness
We have missing data in bmi and chl (not age).
bmi is our outcome, and it will be modeled by age and the complete (missing data imputed) chl variable, as well as their interaction
The missing data in chl will be imputed via a model with age as its predictor!
We're basically fitting two models at once.
Compare
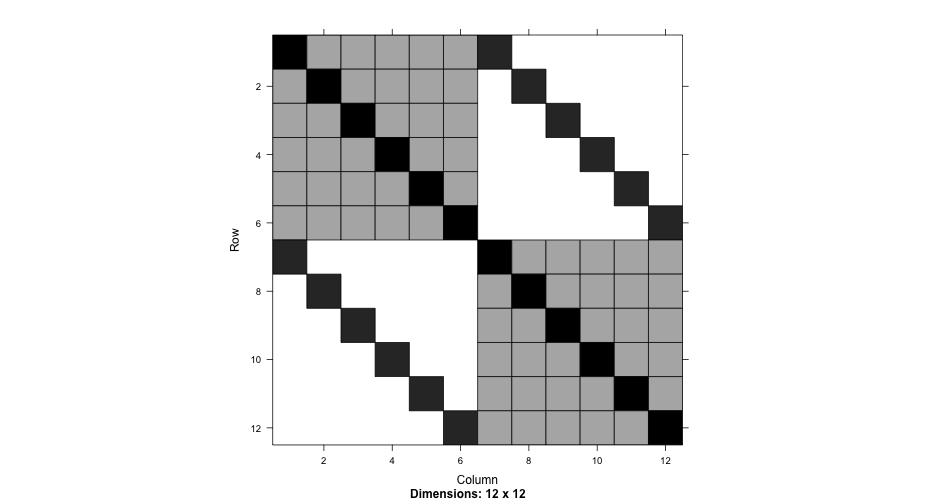
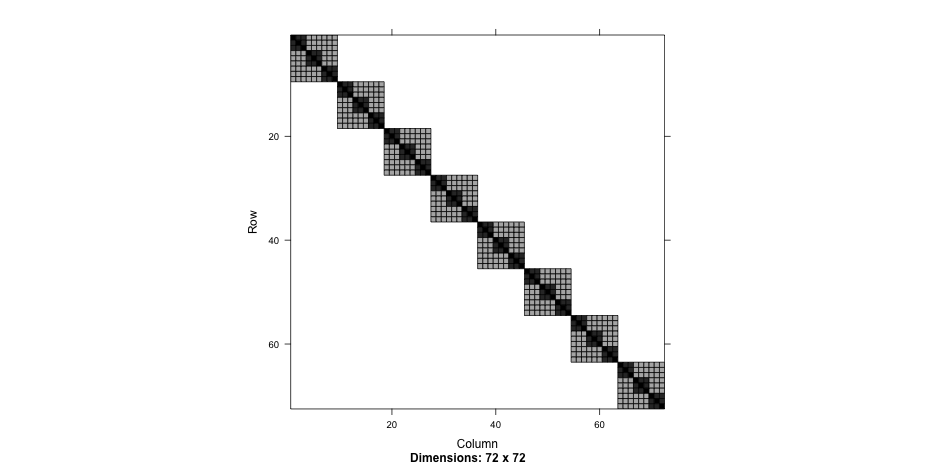
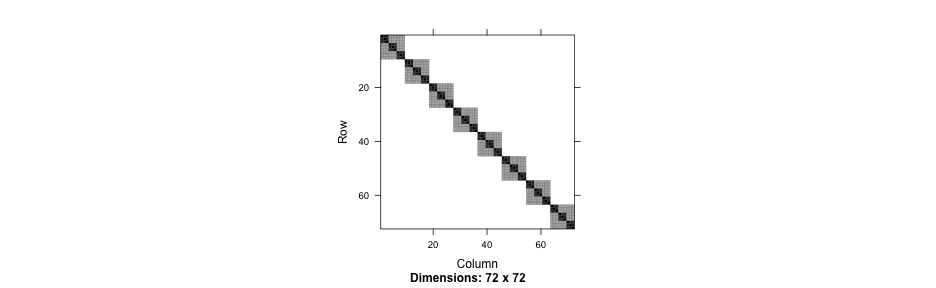
Nested syntax
summary(nested2)## Linear mixed model fit by REML ['lmerMod']## Formula: Thickness ~ 1 + (1 | Lot/Wafer)## Data: Oxide## ## REML criterion at convergence: 454## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -1.8746 -0.4991 0.1047 0.5510 1.7922 ## ## Random effects:## Groups Name Variance Std.Dev.## Wafer:Lot (Intercept) 35.87 5.989 ## Lot (Intercept) 129.91 11.398 ## Residual 12.57 3.545 ## Number of obs: 72, groups: Wafer:Lot, 24; Lot, 8## ## Fixed effects:## Estimate Std. Error t value## (Intercept) 2000.153 4.232 472.6Compare
Explicit IDs
summary(nested3)## Linear mixed model fit by REML ['lmerMod']## Formula: Thickness ~ 1 + (1 | Lot) + (1 | Wafer)## Data: Oxide## ## REML criterion at convergence: 454## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -1.8746 -0.4991 0.1047 0.5510 1.7922 ## ## Random effects:## Groups Name Variance Std.Dev.## Wafer (Intercept) 35.87 5.989 ## Lot (Intercept) 129.91 11.398 ## Residual 12.57 3.545 ## Number of obs: 72, groups: Wafer, 24; Lot, 8## ## Fixed effects:## Estimate Std. Error t value## (Intercept) 2000.153 4.232 472.6The data
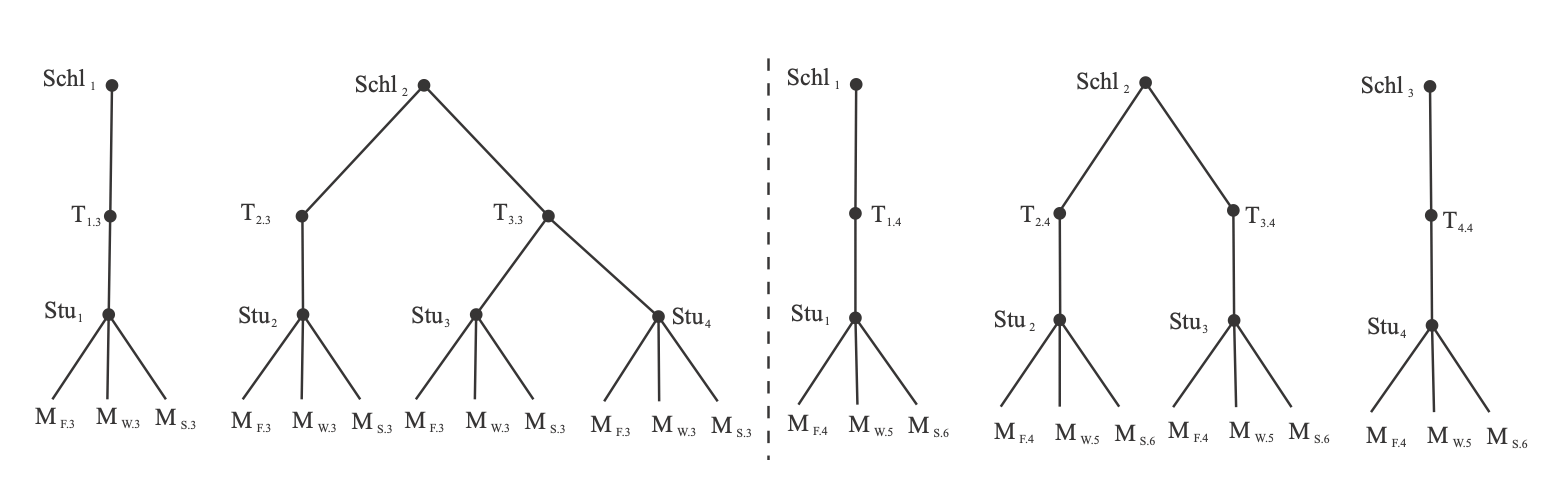
Primary and secondary schools
achievement <- read_csv(here::here("data", "pupils.csv"))achievement## # A tibble: 1,000 x 8## PUPIL primary_school_id secondary_school_id achievement sex ses primary_denominational secondary_denominational## <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> ## 1 1 1 2 6.6 female highest no no ## 2 2 1 1 5.7 male lowest no yes ## 3 3 1 17 4.5 male 2 no no ## 4 4 1 3 4.4 male 2 no no ## 5 5 1 4 5.8 male 3 no yes ## 6 6 1 4 5 female 4 no yes ## 7 7 1 4 4.9 male 3 no yes ## 8 8 1 17 5.3 female 2 no no ## 9 9 1 14 9 male highest no yes ## 10 10 1 22 5.4 male lowest no no ## # … with 990 more rowsccrem <- lmer(achievement ~ 1 + (1|primary_school_id) + (1|secondary_school_id), data = achievement)arm::display(ccrem)## lmer(formula = achievement ~ 1 + (1 | primary_school_id) + (1 | ## secondary_school_id), data = achievement)## coef.est coef.se ## 6.35 0.08 ## ## Error terms:## Groups Name Std.Dev.## primary_school_id (Intercept) 0.41 ## secondary_school_id (Intercept) 0.26 ## Residual 0.72 ## ---## number of obs: 1000, groups: primary_school_id, 50; secondary_school_id, 30## AIC = 2329.1, DIC = 2314.6## deviance = 2317.8Random effects
Note that each random effect would be added to each person's score. This is a little complicated.
str(ranef(ccrem))## List of 2## $ primary_school_id :'data.frame': 50 obs. of 1 variable:## ..$ (Intercept): num [1:50] -0.38 0.209 0.528 0.523 0.312 ...## ..- attr(*, "postVar")= num [1, 1, 1:50] 0.0264 0.0241 0.026 0.023 0.0304 ...## $ secondary_school_id:'data.frame': 30 obs. of 1 variable:## ..$ (Intercept): num [1:30] -0.3854 0.0802 -0.0141 -0.1074 0.1071 ...## ..- attr(*, "postVar")= num [1, 1, 1:30] 0.0165 0.0148 0.0169 0.0139 0.0151 ...## - attr(*, "class")= chr "ranef.mer"Random proportion variables
set.seed(42)achievement <- achievement %>% mutate(primary_school_time = abs(rnorm(1000)), secondary_school_time = abs(rnorm(1000))) %>% rowwise() %>% mutate(tot = primary_school_time + secondary_school_time, primary_school_time = primary_school_time / tot, secondary_school_time = secondary_school_time / tot) %>% ungroup()achievement %>% select(PUPIL, ends_with("time"))## # A tibble: 1,000 x 3## PUPIL primary_school_time secondary_school_time## <dbl> <dbl> <dbl>## 1 1 0.3709286 0.6290714 ## 2 2 0.5186330 0.4813670 ## 3 3 0.2722384 0.7277616 ## 4 4 0.6266984 0.3733016 ## 5 5 0.2887215 0.7112785 ## 6 6 0.1508292 0.8491708 ## 7 7 0.9014468 0.09855322## 8 8 0.03131154 0.9686885 ## 9 9 0.7041820 0.2958180 ## 10 10 0.07281342 0.9271866 ## # … with 990 more rowsItems
Women were asked whether they could engage in the following activities alone (1 = yes, 0 = no):
- Item 1: Go to any part of the village/town/city
- Item 2: Go outside the village/town/city
- Item 3: Talk to a man you do not know
- Item 4: Go to a cinema/cultural show
- Item 5: Go shopping
- Item 6: Go to a cooperative/mothers' club/other club
- Item 7: Attend a political meeting
- Item 8: Go to a health centre/hospital
Read in the data
mobility <- read_csv(here::here("data", "mobility.csv"))mobility## # A tibble: 8,445 x 9## id `Item 1` `Item 2` `Item 3` `Item 4` `Item 5` `Item 6` `Item 7` `Item 8`## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 1 1 1 1 0 0 0 0## 2 2 0 0 0 0 0 0 0 0## 3 3 0 0 0 0 0 0 0 0## 4 4 0 0 0 0 0 0 0 0## 5 5 0 0 0 0 0 0 0 0## 6 6 0 0 0 0 0 0 0 0## 7 7 1 1 0 0 0 0 0 0## 8 8 0 0 0 0 0 0 0 0## 9 9 0 0 0 0 0 0 0 0## 10 10 1 0 0 0 0 0 0 0## # … with 8,435 more rowsmobility_l <- mobility %>% pivot_longer(-id, names_to = "item", values_to = "response")mobility_l## # A tibble: 67,560 x 3## id item response## <dbl> <chr> <dbl>## 1 1 Item 1 1## 2 1 Item 2 1## 3 1 Item 3 1## 4 1 Item 4 1## 5 1 Item 5 0## 6 1 Item 6 0## 7 1 Item 7 0## 8 1 Item 8 0## 9 2 Item 1 0## 10 2 Item 2 0## # … with 67,550 more rowsItem estimates
library(ltm)ltm_onepl <- rasch(mobility[ ,-1])tibble( lme4_ests = fixef(onepl), ltm_ests = ltm_onepl$coefficients[ ,1])## # A tibble: 8 x 2## lme4_ests ltm_ests## <dbl> <dbl>## 1 2.547100 2.523787 ## 2 -1.396082 -1.389602 ## 3 2.082769 2.063258 ## 4 -0.9773621 -0.9687520## 5 -4.591452 -4.626544 ## 6 -3.712541 -3.731430 ## 7 -5.069683 -5.105589 ## 8 -4.184125 -4.212841Person estimates
ltm_theta <- factor.scores(ltm_onepl, resp.patterns = mobility[ ,-1])ltm_discrim <- unique(ltm_onepl$coefficients[ ,2])lme4_theta <- ranef(onepl)$idtheta_compare <- tibble( ltm_theta = ltm_discrim*ltm_theta$score.dat$z1, lme4_theta = lme4_theta[ ,1])theta_compare## # A tibble: 8,445 x 2## ltm_theta lme4_theta## <dbl> <dbl>## 1 1.959870 1.942168 ## 2 -3.364090 -3.346008 ## 3 -3.364090 -3.346008 ## 4 -3.364090 -3.346008 ## 5 -3.364090 -3.346008 ## 6 -3.364090 -3.346008 ## 7 -0.4733693 -0.4751684## 8 -3.364090 -3.346008 ## 9 -3.364090 -3.346008 ## 10 -1.879762 -1.878805 ## # … with 8,435 more rows