Data structuring and basic models
Daniel Anderson
Week 2
First, load the data
library(tidyverse)curran <- read_csv(here::here("data", "curran.csv"))curran## # A tibble: 405 x 15## id anti1 anti2 anti3 anti4 read1 read2 read3 read4 kidgen momage kidage## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 22 1 2 NA NA 2.1 3.9 NA NA 0 28 6.08## 2 34 3 6 4 5 2.1 2.9 4.5 4.5 1 28 6.83## 3 58 0 2 0 1 2.3 4.5 4.2 4.6 0 28 6.5 ## 4 122 0 3 1 1 3.7 8 NA NA 1 28 7.83## 5 125 1 1 2 1 2.3 3.8 4.3 6.2 0 29 7.42## 6 133 3 4 3 5 1.8 2.6 4.1 4 1 28 6.75## 7 163 5 4 5 5 3.5 4.8 5.8 7.5 1 28 7.17## 8 190 0 NA NA 0 2.9 6.1 NA NA 0 28 6.67## 9 227 0 0 2 1 1.8 3.8 4 NA 0 29 6.25## 10 248 1 2 2 0 3.5 5.7 7 6.9 0 28 7.5 ## # … with 395 more rows, and 3 more variables: homecog <dbl>, homeemo <dbl>,## # nmis <dbl>About the data
The data are a sample of 405 children who were within the first two years of entry to elementary school. The data consist of four repeated measures of both the child’s antisocial behavior and the child’s reading recognition skills. In addition, on the first measurement occasion, measures were collected of emotional support and cognitive stimulation provided by the mother. The data were collected using face-to-face interviews of both the child and the mother at two-year intervals between 1986 and 1992.
See here
Let's start easy
First, let's select just the ID variable and the reading scores
read <- curran %>% select(id, starts_with("read"))read## # A tibble: 405 x 5## id read1 read2 read3 read4## <dbl> <dbl> <dbl> <dbl> <dbl>## 1 22 2.1 3.9 NA NA ## 2 34 2.1 2.9 4.5 4.5## 3 58 2.3 4.5 4.2 4.6## 4 122 3.7 8 NA NA ## 5 125 2.3 3.8 4.3 6.2## 6 133 1.8 2.6 4.1 4 ## 7 163 3.5 4.8 5.8 7.5## 8 190 2.9 6.1 NA NA ## 9 227 1.8 3.8 4 NA ## 10 248 3.5 5.7 7 6.9## # … with 395 more rowsMoving to longer
read %>% pivot_longer(cols = read1:read4, names_to = "timepoint", values_to = "score")## # A tibble: 1,620 x 3## id timepoint score## <dbl> <chr> <dbl>## 1 22 read1 2.1## 2 22 read2 3.9## 3 22 read3 NA ## 4 22 read4 NA ## 5 34 read1 2.1## 6 34 read2 2.9## 7 34 read3 4.5## 8 34 read4 4.5## 9 58 read1 2.3## 10 58 read2 4.5## # … with 1,610 more rowsAlternative
You can also specify the columns that should not be pivoted
read %>% pivot_longer(-id, names_to = "timepoint", values_to = "score")## # A tibble: 1,620 x 3## id timepoint score## <dbl> <chr> <dbl>## 1 22 read1 2.1## 2 22 read2 3.9## 3 22 read3 NA ## 4 22 read4 NA ## 5 34 read1 2.1## 6 34 read2 2.9## 7 34 read3 4.5## 8 34 read4 4.5## 9 58 read1 2.3## 10 58 read2 4.5## # … with 1,610 more rowsMutate
- Use
mutate()to modify the column afterwords
Why did I subtract 1?
read %>% pivot_longer(-id, names_to = "timepoint", values_to = "score") %>% mutate(timepoint = parse_number(timepoint) - 1)## # A tibble: 1,620 x 3## id timepoint score## <dbl> <dbl> <dbl>## 1 22 0 2.1## 2 22 1 3.9## 3 22 2 NA ## 4 22 3 NA ## 5 34 0 2.1## 6 34 1 2.9## 7 34 2 4.5## 8 34 3 4.5## 9 58 0 2.3## 10 58 1 4.5## # … with 1,610 more rowsTransform during the pivot
read %>% pivot_longer(-id, names_to = "timepoint", values_to = "score", names_transform = list( timepoint = parse_number) )## # A tibble: 1,620 x 3## id timepoint score## <dbl> <dbl> <dbl>## 1 22 1 2.1## 2 22 2 3.9## 3 22 3 NA ## 4 22 4 NA ## 5 34 1 2.1## 6 34 2 2.9## 7 34 3 4.5## 8 34 4 4.5## 9 58 1 2.3## 10 58 2 4.5## # … with 1,610 more rowsAlternative transformation
This does the subtraction by 1 also
sub1 <- function(x) parse_number(x) - 1read %>% pivot_longer(-id, names_to = "timepoint", values_to = "score", names_transform = list(timepoint = sub1))## # A tibble: 1,620 x 3## id timepoint score## <dbl> <dbl> <dbl>## 1 22 0 2.1## 2 22 1 3.9## 3 22 2 NA ## 4 22 3 NA ## 5 34 0 2.1## 6 34 1 2.9## 7 34 2 4.5## 8 34 3 4.5## 9 58 0 2.3## 10 58 1 4.5## # … with 1,610 more rowsYet another approach
This one doesn't subtract 1, however
read %>% pivot_longer(-id, names_to = "timepoint", values_to = "score", names_prefix = "read", names_ptype = list(timepoint = "numeric"))## # A tibble: 1,620 x 3## id timepoint score## <dbl> <chr> <dbl>## 1 22 1 2.1## 2 22 2 3.9## 3 22 3 NA ## 4 22 4 NA ## 5 34 1 2.1## 6 34 2 2.9## 7 34 3 4.5## 8 34 4 4.5## 9 58 1 2.3## 10 58 2 4.5## # … with 1,610 more rowsMoving back
Although moving longer is most often useful for multilevel modeling, occasionally we need to go wider - e.g., for a join.
First, let's create a longer data object
l <- read %>% pivot_longer(-id, names_to = "timepoint", values_to = "score", names_transform = list(timepoint = sub1))Now let's move it back
Use pivot_wider() instead
l %>% pivot_wider(names_from = timepoint, values_from = score)## # A tibble: 405 x 5## id `0` `1` `2` `3`## <dbl> <dbl> <dbl> <dbl> <dbl>## 1 22 2.1 3.9 NA NA ## 2 34 2.1 2.9 4.5 4.5## 3 58 2.3 4.5 4.2 4.6## 4 122 3.7 8 NA NA ## 5 125 2.3 3.8 4.3 6.2## 6 133 1.8 2.6 4.1 4 ## 7 163 3.5 4.8 5.8 7.5## 8 190 2.9 6.1 NA NA ## 9 227 1.8 3.8 4 NA ## 10 248 3.5 5.7 7 6.9## # … with 395 more rowsChallenge
Let's go back to the full curran data. See if you can get your data to look like the below.
There are, again, multiple ways to do this, including only through pivot_longer
## # A tibble: 3,240 x 10## id kidgen momage kidage homecog homeemo nmis variable timepoint value## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>## 1 22 0 28 6.08 13 10 4 anti 0 1 ## 2 22 0 28 6.08 13 10 4 anti 1 2 ## 3 22 0 28 6.08 13 10 4 anti 2 NA ## 4 22 0 28 6.08 13 10 4 anti 3 NA ## 5 22 0 28 6.08 13 10 4 read 0 2.1## 6 22 0 28 6.08 13 10 4 read 1 3.9## 7 22 0 28 6.08 13 10 4 read 2 NA ## 8 22 0 28 6.08 13 10 4 read 3 NA ## 9 34 1 28 6.83 9 9 0 anti 0 3 ## 10 34 1 28 6.83 9 9 0 anti 1 6 ## # … with 3,230 more rows06:00
More transforming
Our data is probably still not in the format we want. Can you get it in the format like the below?
## # A tibble: 1,620 x 10## id kidgen momage kidage homecog homeemo nmis timepoint anti read## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 22 0 28 6.08 13 10 4 0 1 2.1## 2 22 0 28 6.08 13 10 4 1 2 3.9## 3 22 0 28 6.08 13 10 4 2 NA NA ## 4 22 0 28 6.08 13 10 4 3 NA NA ## 5 34 1 28 6.83 9 9 0 0 3 2.1## 6 34 1 28 6.83 9 9 0 1 6 2.9## 7 34 1 28 6.83 9 9 0 2 4 4.5## 8 34 1 28 6.83 9 9 0 3 5 4.5## 9 58 0 28 6.5 9 6 0 0 0 2.3## 10 58 0 28 6.5 9 6 0 1 2 4.5## # … with 1,610 more rows04:00
Another example
Read in the letter sounds data
ls <- read_csv(here::here("data", "ls19.csv"))ls## # A tibble: 962 x 13## county distid dist_name instid inst_name inst_type## <chr> <dbl> <chr> <dbl> <chr> <chr> ## 1 All Counties 9999 Statewide 9999 Statewide State ## 2 Baker 1894 Baker SD 5J 1894 Baker SD 5J District ## 3 Baker 1895 Huntington SD 16J 1895 Huntington SD 16J District ## 4 Baker 1896 Burnt River SD 30J 1896 Burnt River SD 30J District ## 5 Baker 1897 Pine Eagle SD 61 1897 Pine Eagle SD 61 District ## 6 Benton 1898 Monroe SD 1J 1898 Monroe SD 1J District ## 7 Benton 1899 Alsea SD 7J 1899 Alsea SD 7J District ## 8 Benton 1900 Philomath SD 17J 1900 Philomath SD 17J District ## 9 Benton 1901 Corvallis SD 509J 1901 Corvallis SD 509J District ## 10 Clackamas 1902 Clackamas ESD 1902 Clackamas ESD District ## # … with 952 more rows, and 7 more variables: asian <dbl>,## # black_african_american <dbl>, hispanic_latino <dbl>,## # american_indian_alaska_native <dbl>, multi_racial <dbl>,## # native_hawaiian_pacific_islander <dbl>, white <dbl>LS Data
Average scores on the letter sounds portion of the kindergarten entry assessment for every school in the state, by race.
Data missing if n too small
Remember - you (generally) don't need to dummy-code variables in R
Try structuring this data so you could estimate between-district variability, while accounting for race/ethnicity
06:00
Self-regulation data
Same basic data with a different outcome and a different structure.
Try restructuring this one
selfreg <- read_csv(here::here("data", "selfreg19.csv"))selfreg## # A tibble: 6,734 x 13## county distid dist_name instid inst_name inst_type selfreg_score## <chr> <dbl> <chr> <dbl> <chr> <chr> <dbl>## 1 All Counties 9999 Statewide 9999 Statewide State 3.7## 2 All Counties 9999 Statewide 9999 Statewide State 3.3## 3 All Counties 9999 Statewide 9999 Statewide State 3.4## 4 All Counties 9999 Statewide 9999 Statewide State 3.3## 5 All Counties 9999 Statewide 9999 Statewide State 3.5## 6 All Counties 9999 Statewide 9999 Statewide State 3.4## 7 All Counties 9999 Statewide 9999 Statewide State 3.5## 8 Baker 1894 Baker SD 5J 1894 Baker SD 5J District NA ## 9 Baker 1894 Baker SD 5J 1894 Baker SD 5J District NA ## 10 Baker 1894 Baker SD 5J 1894 Baker SD 5J District 3.9## # … with 6,724 more rows, and 6 more variables: Asian <dbl>,## # Black.African.American <dbl>, Hispanic.Latino <dbl>, Multi.Racial <dbl>,## # Native.Hawaiian.Pacific.Islander <dbl>, White <dbl>06:00
A bit of a caveat
The preceding examples would lead to sort of fundamentally flawed analyses
We'd be estimating each district mean as the mean of the school means
There are ways to account for this, which we may or may not get into later in the term
Could potentially try weighting each school mean by the school size
Model fitting
We could start with a fully unconditional model (not unconditional growth), but that's really a misspecification in this case - we know we have to account for time.
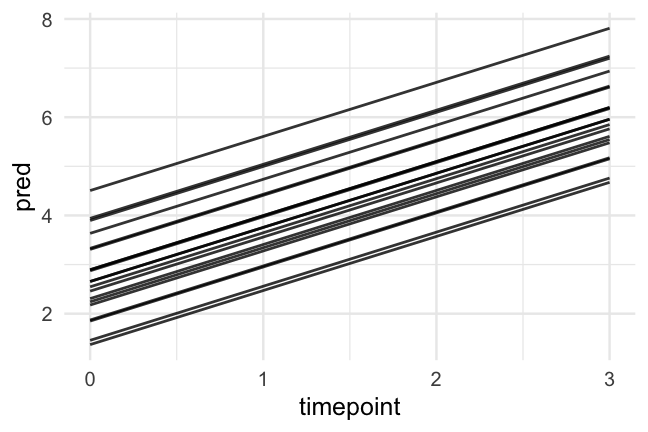
Let's first fit a model with random intercepts
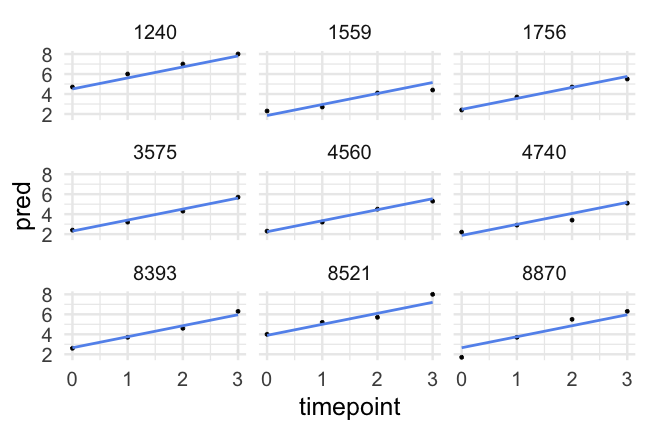
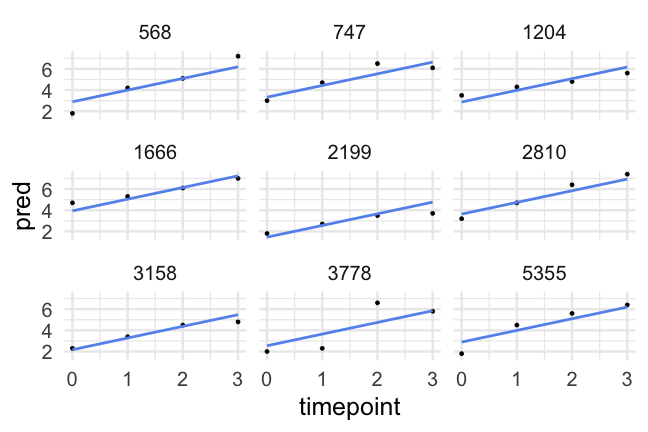
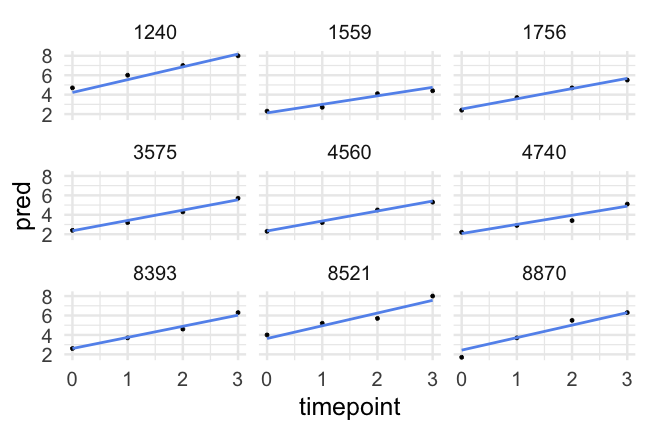
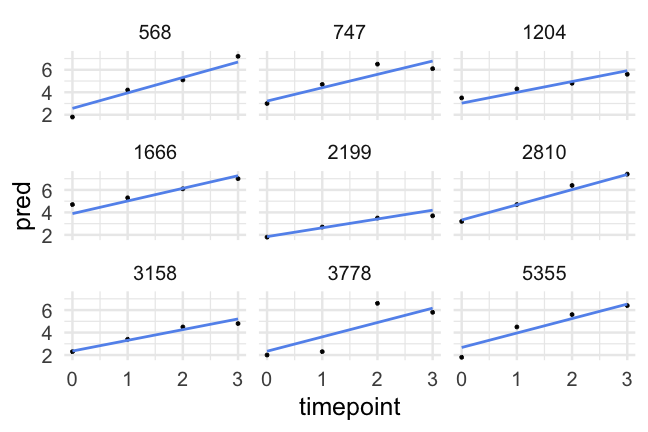
- A reminder of what the data look like
d## # A tibble: 1,620 x 10## id kidgen momage kidage homecog homeemo nmis timepoint anti read## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 22 0 28 6.08 13 10 4 0 1 2.1## 2 22 0 28 6.08 13 10 4 1 2 3.9## 3 22 0 28 6.08 13 10 4 2 NA NA ## 4 22 0 28 6.08 13 10 4 3 NA NA ## 5 34 1 28 6.83 9 9 0 0 3 2.1## 6 34 1 28 6.83 9 9 0 1 6 2.9## 7 34 1 28 6.83 9 9 0 2 4 4.5## 8 34 1 28 6.83 9 9 0 3 5 4.5## 9 58 0 28 6.5 9 6 0 0 0 2.3## 10 58 0 28 6.5 9 6 0 1 2 4.5## # … with 1,610 more rowsParallel slopes
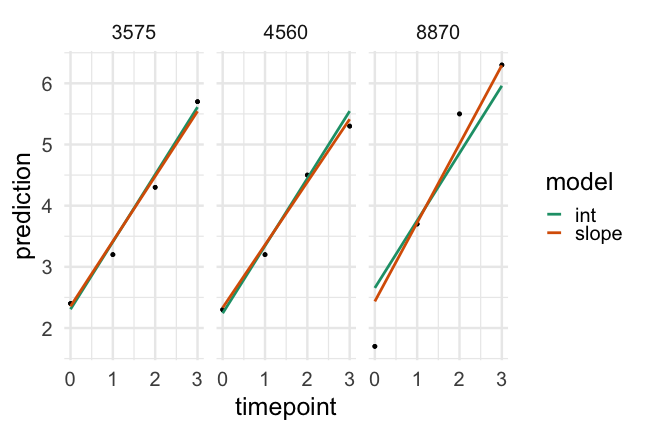
Had we of fit a standard regression model we would have had one slope to represent the trend of all participants, which would (fairly clearly) be less than ideal
Now, we've allowed each participant to have a different starting point, but constrained the rate of change to be constant.
How reasonable is this assumption?
Model summary
summary(m_intercepts)## Linear mixed model fit by REML ['lmerMod']## Formula: read ~ 1 + timepoint + (1 | id)## Data: d## ## REML criterion at convergence: 3487.6## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -2.6170 -0.5207 0.0383 0.5214 3.7428 ## ## Random effects:## Groups Name Variance Std.Dev.## id (Intercept) 0.7797 0.8830 ## Residual 0.4609 0.6789 ## Number of obs: 1325, groups: id, 405## ## Fixed effects:## Estimate Std. Error t value## (Intercept) 2.70374 0.05257 51.43## timepoint 1.10134 0.01759 62.62## ## Correlation of Fixed Effects:## (Intr)## timepoint -0.406Modeling
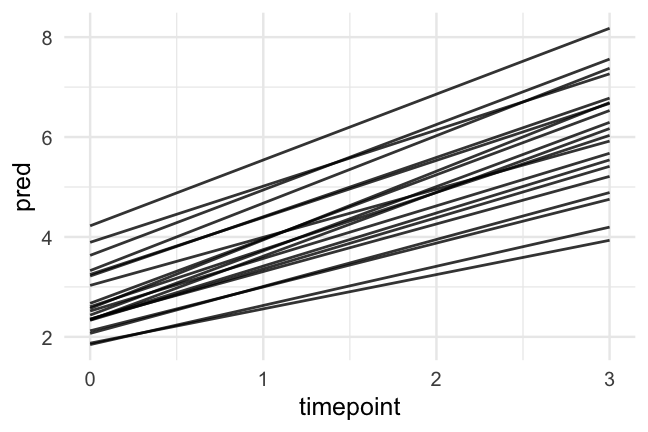
- Let's fit a second model that allows each participant to have a different slope
m_slopes <- lmer(read ~ 1 + timepoint + (1 + timepoint|id), data = d)Quick note on syntax
I'm being very explicit in the above about what I'm estimating. However, intercepts are generally implied. So the above is equivalent to
m_slopes <- lmer(read ~ timepoint + (timepoint|id), data = d)which is actually how I generally write it
What's the output look like
summary(m_slopes)## Linear mixed model fit by REML ['lmerMod']## Formula: read ~ 1 + timepoint + (1 + timepoint | id)## Data: d## ## REML criterion at convergence: 3382## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -2.7161 -0.5201 -0.0220 0.4793 4.1847 ## ## Random effects:## Groups Name Variance Std.Dev. Corr## id (Intercept) 0.57309 0.7570 ## timepoint 0.07459 0.2731 0.29## Residual 0.34584 0.5881 ## Number of obs: 1325, groups: id, 405## ## Fixed effects:## Estimate Std. Error t value## (Intercept) 2.69609 0.04530 59.52## timepoint 1.11915 0.02169 51.60## ## Correlation of Fixed Effects:## (Intr)## timepoint -0.155P values
The {lme4} package does not report p-values. This is because its author, Douglas Bates, believes they are fundamentally flawed for multilevel models.
The link above is worth reading through, but basically it is not straightforward to calculate the denominator degrees of freedom for an F test. The methods that are used are approximations and, although generally accepted, are not guaranteed to be correct.
Alternatives
There are two primary work-arounds here:
Don't use p-values, and instead just interpret the confidence intervals, or
Use the same approximation that others use via {lmerTest} package.
Confidence intervals
Multiple options, but profiled or bootstrap confidence intervals are generally preferred, though computationally intensive. Note that these provide CIs for the variance components as well.
confint(m_slopes)## Computing profile confidence intervals ...## 2.5 % 97.5 %## .sig01 0.67961051 0.8365439## .sig02 0.06898955 0.5434982## .sig03 0.22282787 0.3213734## .sigma 0.55548063 0.6238545## (Intercept) 2.60721096 2.7850364## timepoint 1.07653165 1.1622218New summary
summary(m_slopes2)## Linear mixed model fit by REML. t-tests use Satterthwaite's method [## lmerModLmerTest]## Formula: read ~ timepoint + (timepoint | id)## Data: d## ## REML criterion at convergence: 3382## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -2.7161 -0.5201 -0.0220 0.4793 4.1847 ## ## Random effects:## Groups Name Variance Std.Dev. Corr## id (Intercept) 0.57309 0.7570 ## timepoint 0.07459 0.2731 0.29## Residual 0.34584 0.5881 ## Number of obs: 1325, groups: id, 405## ## Fixed effects:## Estimate Std. Error df t value Pr(>|t|) ## (Intercept) 2.69609 0.04530 400.87693 59.52 <2e-16 ***## timepoint 1.11915 0.02169 308.40833 51.60 <2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## ## Correlation of Fixed Effects:## (Intr)## timepoint -0.155Using built-in approaches
anova(m_intercepts, m_slopes)## refitting model(s) with ML (instead of REML)## Data: d## Models:## m_intercepts: read ~ 1 + timepoint + (1 | id)## m_slopes: read ~ 1 + timepoint + (1 + timepoint | id)## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) ## m_intercepts 4 3485.1 3505.8 -1738.5 3477.1 ## m_slopes 6 3383.8 3414.9 -1685.9 3371.8 105.29 2 < 2.2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1What does this mean?
The {performance} package
Similar information, little bit nicer output
library(performance)compare_performance(m_intercepts, m_slopes) %>% print_md()Table: Comparison of Model Performance Indices
| Name | Model | AIC | BIC | R2 (cond.) | R2 (marg.) | ICC | RMSE | Sigma |
|---|---|---|---|---|---|---|---|---|
| m_intercepts | lmerMod | 3495.56 | 3516.32 | 0.83 | 0.55 | 0.63 | 0.59 | 0.68 |
| m_slopes | lmerMod | 3394.00 | 3425.14 | 0.88 | 0.54 | 0.73 | 0.47 | 0.59 |
Quick note on Bayes factors
Pure Bayesians typically hate them - they are sometimes called a Bayesain p-value
Tests under which model the observed data are more likely
Larger values indicate less support for the comparison model
I would advise you only use it in combination with other sources of evidence
See here for more information
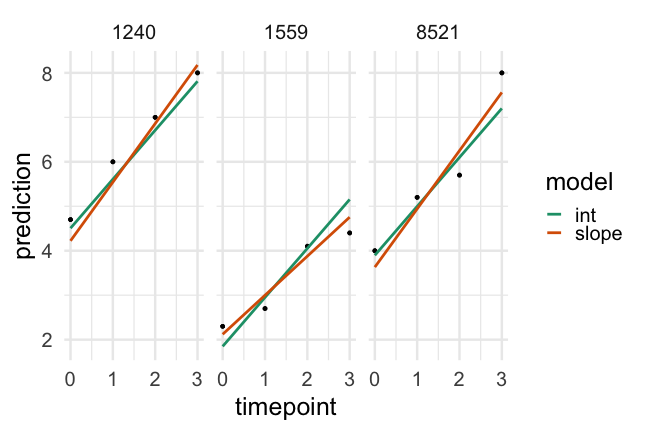
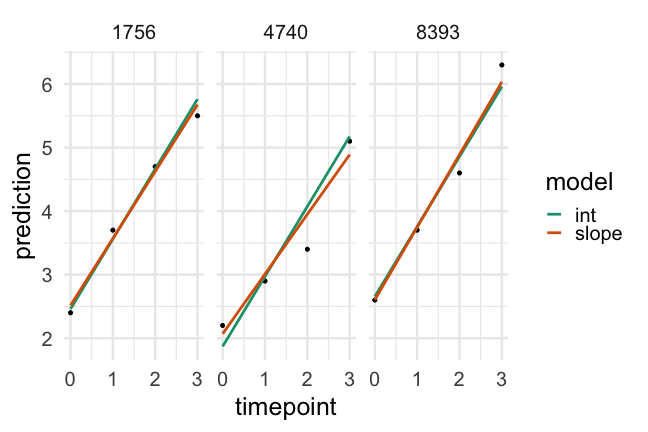
Conclusions
Given the evidence we've looked at I would conclude:
- Both models are a considerable improvement over a linear regression model
- The random intercepts and slopes model is a better fit to the data than the random intercepts only model
- There is more variability in initial starting point than rate of change (which is typical)