Predictions and visualizations
Daniel Anderson
Week 3
Learning objectives
Understand how to pull different pieces out of the model
Understand how multilevel models make their predictions for individual observations
- And specifically how they differ from single-level regression models
Be able to use the output from model objects to visualize different parts of the model.
Read in the data
Let's read in the popularity data so we can fit some really basic models.
You try first
02:00
library(tidyverse)popular <- read_csv(here::here("data", "popularity.csv"))popular## # A tibble: 2,000 x 7## pupil class extrav sex texp popular popteach## <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>## 1 1 1 5 girl 24 6.3 6## 2 2 1 7 boy 24 4.9 5## 3 3 1 4 girl 24 5.3 6## 4 4 1 3 girl 24 4.7 5## 5 5 1 5 girl 24 6 6## 6 6 1 4 boy 24 4.7 5## 7 7 1 5 boy 24 5.9 5## 8 8 1 4 boy 24 4.2 5## 9 9 1 5 boy 24 5.2 5## 10 10 1 5 boy 24 3.9 3## # … with 1,990 more rowsFit a basic model
Fit each of the following models
popularas the outcome, with a random intercept forclasspopularas the outcome, withsexincluded as a fixed effect and a random intercept forclasspopularas the outcome, withsexincluded as a fixed effect and a random intercept and slope forclass
04:00
Compare performance
Use whatever tests you'd like, and come up with the model you think fits the data best
04:00
library(performance)compare_performance(m0, m1, m2) %>% print_md()Table: Comparison of Model Performance Indices
| Name | Model | AIC | BIC | R2 (cond.) | R2 (marg.) | ICC | RMSE | Sigma |
|---|---|---|---|---|---|---|---|---|
| m0 | lmerMod | 6336.51 | 6353.31 | 0.36 | 0.00 | 0.36 | 1.08 | 1.11 |
| m1 | lmerMod | 5572.07 | 5594.48 | 0.53 | 0.26 | 0.37 | 0.89 | 0.91 |
| m2 | lmerMod | 5571.05 | 5604.66 | 0.54 | 0.26 | 0.38 | 0.88 | 0.90 |
Test of the Log-likelihood
test_likelihoodratio(m0, m1) %>% print_md()| Name | Model | df | df_diff | Chi2 | p |
|---|---|---|---|---|---|
| m0 | lmerMod | 3 | |||
| m1 | lmerMod | 4 | 1 | 766.44 | 1.07e-168 |
test_likelihoodratio(m1, m2) %>% print_md()| Name | Model | df | df_diff | Chi2 | p |
|---|---|---|---|---|---|
| m1 | lmerMod | 4 | |||
| m2 | lmerMod | 6 | 2 | 5.02 | 0.08 |
Pretty good evidence that m1 displays the best fit
Package options
The {parameters} package (part of the {easystats} ecosystem) will get you where you want to be.
Alternatively, you could use the {broom.mixed} package (which is a spin-off of the {broom} package) which requires slightly less code.
I'll illustrate both
install.packages("broom.mixed"){broom.mixed}
library(broom.mixed)tidy(m0)## # A tibble: 3 x 6## effect group term estimate std.error statistic## <chr> <chr> <chr> <dbl> <dbl> <dbl>## 1 fixed <NA> (Intercept) 5.077860 0.08739443 58.10278## 2 ran_pars class sd__(Intercept) 0.8379169 NA NA ## 3 ran_pars Residual sd__Observation 1.105348 NA NA{broom.mixed}
library(broom.mixed)tidy(m0)## # A tibble: 3 x 6## effect group term estimate std.error statistic## <chr> <chr> <chr> <dbl> <dbl> <dbl>## 1 fixed <NA> (Intercept) 5.077860 0.08739443 58.10278## 2 ran_pars class sd__(Intercept) 0.8379169 NA NA ## 3 ran_pars Residual sd__Observation 1.105348 NA NAOr get just the fixed effects
tidy(m0, effects = "fixed")## # A tibble: 1 x 5## effect term estimate std.error statistic## <chr> <chr> <dbl> <dbl> <dbl>## 1 fixed (Intercept) 5.077860 0.08739443 58.10278Across models
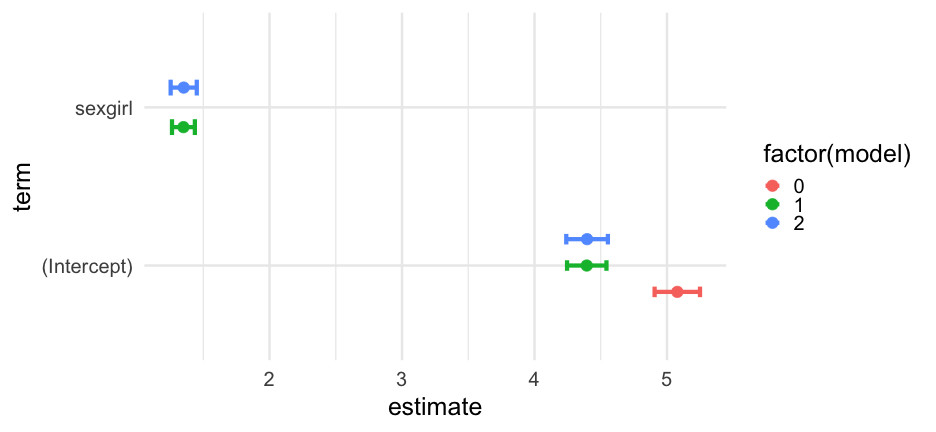
Let's tidy all three models, extracting just the fixed effects, and adding in a 95% confidence interval
models <- bind_rows( tidy(m0, effects = "fixed", conf.int = TRUE), tidy(m1, effects = "fixed", conf.int = TRUE), tidy(m2, effects = "fixed", conf.int = TRUE), .id = "model" ) %>% mutate(model = as.numeric(model) - 1)models## # A tibble: 5 x 8## model effect term estimate std.error statistic conf.low conf.high## <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 0 fixed (Intercept) 5.077860 0.08739443 58.10278 4.906570 5.249150## 2 1 fixed (Intercept) 4.394460 0.07586494 57.92478 4.245767 4.543152## 3 1 fixed sexgirl 1.350102 0.04403301 30.66113 1.263799 1.436405## 4 2 fixed (Intercept) 4.396820 0.08012978 54.87123 4.239768 4.553871## 5 2 fixed sexgirl 1.352175 0.05029920 26.88263 1.253590 1.450760Standardization
The nice thing about tidying the model output, is that the code on the previous slides will always work regardless of the model(s) you've fit.
Caveat
I don't actually think the prior plot is all that useful.
Occasionally helpful to visualize differences between coefficients, but you'll usually want to omit the intercept
Be careful about scales
If you were to publish the prior plot, make it prettier and more accessible first
{parameters}
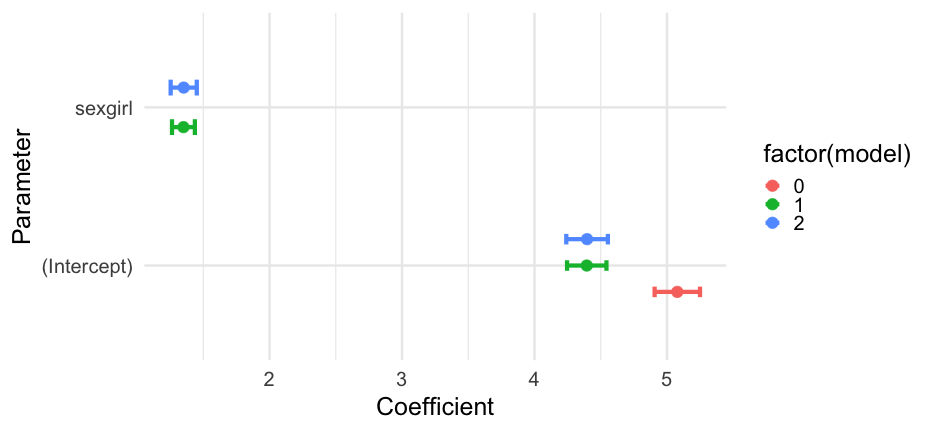
To get basically the same output from parameters:
library(parameters)parameters(m0) %>% as_tibble()## # A tibble: 1 x 10## Parameter Coefficient SE CI CI_low CI_high t df_error## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int>## 1 (Intercept) 5.077860 0.08739443 0.95 4.906570 5.249150 58.10278 1997## # … with 2 more variables: p <dbl>, Effects <chr>models2 <- bind_rows( as_tibble(parameters(m0)), as_tibble(parameters(m1)), as_tibble(parameters(m2)), .id = "model" ) %>% mutate(model = as.numeric(model) - 1)Other parts of the model
From here on out, I'll just be using {broom.mixed} but either package should work, with minor tweaks.
Variance components
Notice that, by default, we don't have any uncertainty
tidy(m0)## # A tibble: 3 x 6## effect group term estimate std.error statistic## <chr> <chr> <chr> <dbl> <dbl> <dbl>## 1 fixed <NA> (Intercept) 5.077860 0.08739443 58.10278## 2 ran_pars class sd__(Intercept) 0.8379169 NA NA ## 3 ran_pars Residual sd__Observation 1.105348 NA NAOther parts of the model
From here on out, I'll just be using {broom.mixed} but either package should work, with minor tweaks.
Variance components
Notice that, by default, we don't have any uncertainty
tidy(m0)## # A tibble: 3 x 6## effect group term estimate std.error statistic## <chr> <chr> <chr> <dbl> <dbl> <dbl>## 1 fixed <NA> (Intercept) 5.077860 0.08739443 58.10278## 2 ran_pars class sd__(Intercept) 0.8379169 NA NA ## 3 ran_pars Residual sd__Observation 1.105348 NA NAWe can fix this by using bootstrap or profiled CIs
Bootstrap CIs
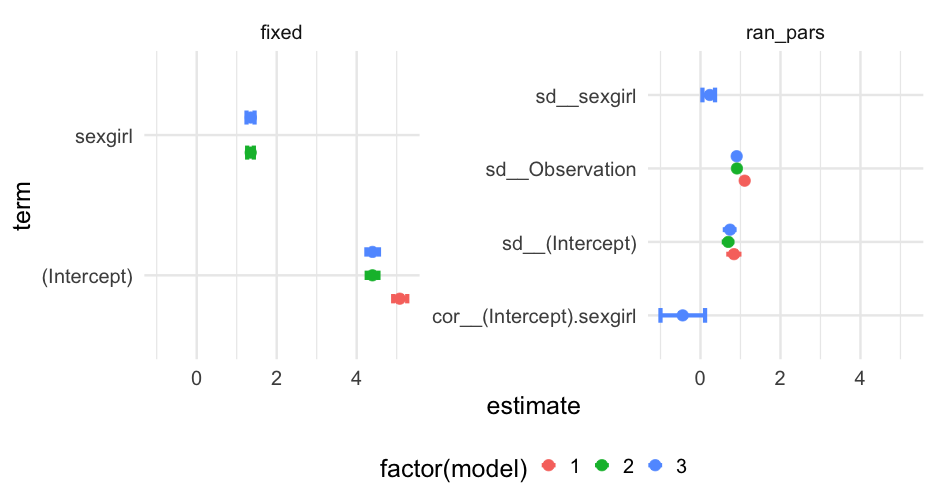
tidy( m0, effects = "ran_pars", conf.int = TRUE, conf.method = "boot")## Computing bootstrap confidence intervals ...## # A tibble: 2 x 6## effect group term estimate conf.low conf.high## <chr> <chr> <chr> <dbl> <dbl> <dbl>## 1 ran_pars class sd__(Intercept) 0.8379169 0.7145176 0.9652536## 2 ran_pars Residual sd__Observation 1.105348 1.071117 1.141232m2
Note this takes a bit of time, even though the model is pretty simple. Also these are standard deviations and a correlation, not variances/covariances
tidy( m2, effects = "ran_pars", conf.int = TRUE, conf.method = "boot")## Computing bootstrap confidence intervals ...## ## 35 message(s): boundary (singular) fit: see ?isSingular## 16 warning(s): Model failed to converge with max|grad| = 0.00213278 (tol = 0.002, component 1) (and others)## # A tibble: 4 x 6## effect group term estimate conf.low conf.high## <chr> <chr> <chr> <dbl> <dbl> <dbl>## 1 ran_pars class sd__(Intercept) 0.7389520 0.6133643 0.8469480 ## 2 ran_pars class cor__(Intercept).sexgirl -0.4442543 -1 0.04996563## 3 ran_pars class sd__sexgirl 0.2336921 0.04570838 0.3513648 ## 4 ran_pars Residual sd__Observation 0.9049626 0.8744993 0.9341371One approach
pull_model_results <- function(model) { tidy( model, conf.int = TRUE, conf.method = "boot" )}full_models <- bind_rows( pull_model_results(m0), pull_model_results(m1), pull_model_results(m2), .id = "model")## Computing bootstrap confidence intervals ...## Computing bootstrap confidence intervals ...## Computing bootstrap confidence intervals ...## Computing bootstrap confidence intervals ...## Computing bootstrap confidence intervals ...## ## 30 message(s): boundary (singular) fit: see ?isSingular## 13 warning(s): Model failed to converge with max|grad| = 0.00215039 (tol = 0.002, component 1) (and others)## Computing bootstrap confidence intervals ...## ## 30 message(s): boundary (singular) fit: see ?isSingular## 9 warning(s): Model failed to converge with max|grad| = 0.00203702 (tol = 0.002, component 1) (and others)Random effects
Pop Quiz
What's the difference between the output from below and the output on the next slide?
tidy(m0, effects = "ran_vals")## # A tibble: 100 x 6## effect group level term estimate std.error## <chr> <chr> <chr> <chr> <dbl> <dbl>## 1 ran_vals class 1 (Intercept) -0.002630828 0.2370649## 2 ran_vals class 2 (Intercept) -0.8949875 0.2370649## 3 ran_vals class 3 (Intercept) -0.3496155 0.2487845## 4 ran_vals class 4 (Intercept) 0.3318175 0.2222273## 5 ran_vals class 5 (Intercept) 0.1919485 0.2317938## 6 ran_vals class 6 (Intercept) -0.6833977 0.2370649## 7 ran_vals class 7 (Intercept) -0.8062842 0.2317938## 8 ran_vals class 8 (Intercept) -1.019181 0.2370649## 9 ran_vals class 9 (Intercept) -0.3844123 0.2370649## 10 ran_vals class 10 (Intercept) 0.2226622 0.2178678## # … with 90 more rowstidy(m0, effects = "ran_coefs")## # A tibble: 100 x 5## effect group level term estimate## <chr> <chr> <chr> <chr> <dbl>## 1 ran_coefs class 1 (Intercept) 5.075229## 2 ran_coefs class 2 (Intercept) 4.182872## 3 ran_coefs class 3 (Intercept) 4.728244## 4 ran_coefs class 4 (Intercept) 5.409677## 5 ran_coefs class 5 (Intercept) 5.269808## 6 ran_coefs class 6 (Intercept) 4.394462## 7 ran_coefs class 7 (Intercept) 4.271575## 8 ran_coefs class 8 (Intercept) 4.058678## 9 ran_coefs class 9 (Intercept) 4.693447## 10 ran_coefs class 10 (Intercept) 5.300522## # … with 90 more rowsAnswer
The output from ran_vals provides the estimate from αj∼N(0,σ).
The output from ran_coefs provides the class-level predictions, i.e., in this case, the intercept + the estimated ran_vals.
Example
tidy(m0, effects = "ran_vals")$estimate[1:5] + fixef(m0)[1]## [1] 5.075229 4.182872 4.728244 5.409677 5.269808tidy(m0, effects = "ran_coefs")$estimate[1:5]## [1] 5.075229 4.182872 4.728244 5.409677 5.269808Try again
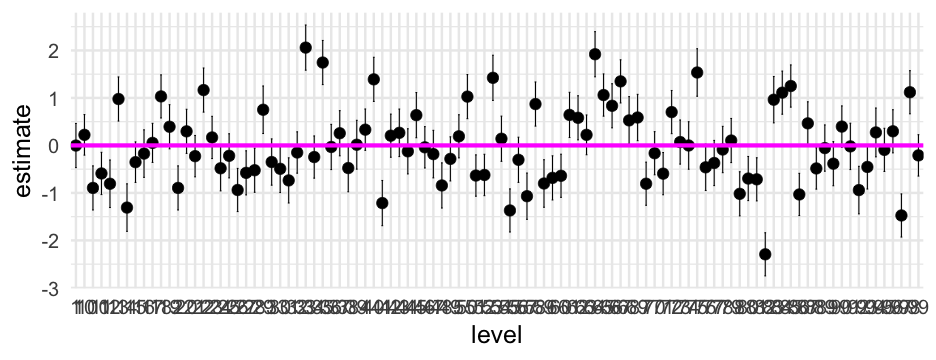
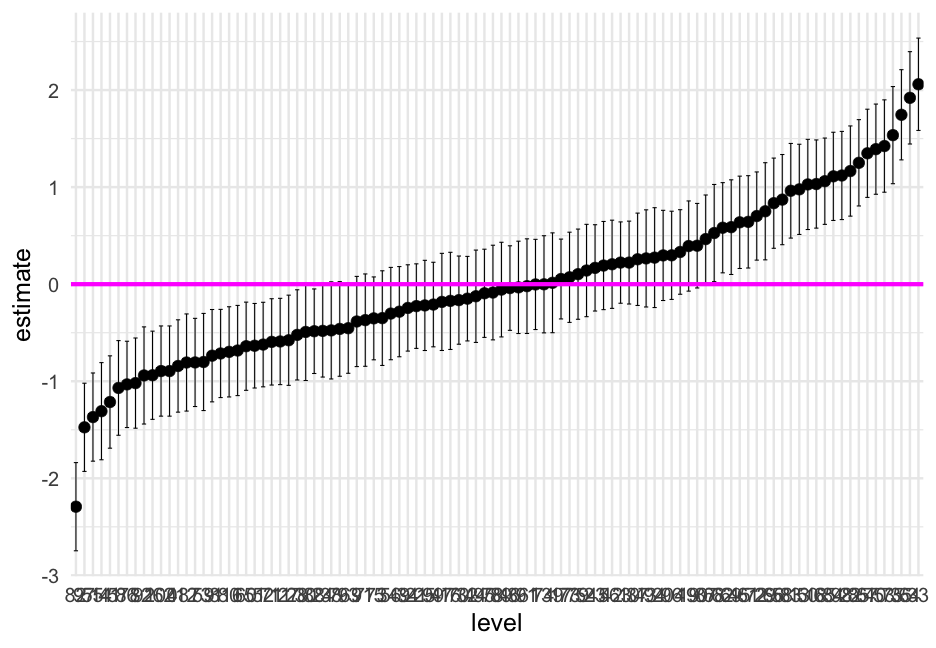
Let's reorder the level according to the estimate
m0_ranvals %>% mutate(level = reorder(factor(level), estimate)) %>% ggplot(aes(level, estimate)) + geom_errorbar(aes(ymin = conf.low, ymax = conf.high), width = 0.5) + geom_point() + geom_hline(yintercept = 0, size = 2, color = "magenta")Wrapping up coef plots
Coefficient plots are generally fairly easy to produce, but often not the most informative
Random effects plots are probs more informative than the fixed effects/variance components plots
You can make either a lot more fancy, accessible, etc., and probably should if you're going to use it for publication.
Reminder
Our raw data looks like this
popular## # A tibble: 2,000 x 7## pupil class extrav sex texp popular popteach## <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>## 1 1 1 5 girl 24 6.3 6## 2 2 1 7 boy 24 4.9 5## 3 3 1 4 girl 24 5.3 6## 4 4 1 3 girl 24 4.7 5## 5 5 1 5 girl 24 6 6## 6 6 1 4 boy 24 4.7 5## 7 7 1 5 boy 24 5.9 5## 8 8 1 4 boy 24 4.2 5## 9 9 1 5 boy 24 5.2 5## 10 10 1 5 boy 24 3.9 3## # … with 1,990 more rowsMaking a prediction
What would our model on the prior slide predict for the first student?
pupil1 <- popular[1, ]pupil1## # A tibble: 1 x 7## pupil class extrav sex texp popular popteach## <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>## 1 1 1 5 girl 24 6.3 6coef(m)[1] + # intercept coef(m)[2] * (pupil1$sex == "girl")## (Intercept) ## 5.853314Making a prediction
What would our model on the prior slide predict for the first student?
pupil1 <- popular[1, ]pupil1## # A tibble: 1 x 7## pupil class extrav sex texp popular popteach## <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>## 1 1 1 5 girl 24 6.3 6coef(m)[1] + # intercept coef(m)[2] * (pupil1$sex == "girl")## (Intercept) ## 5.853314predict(m)[1]## 1 ## 5.853314Thinking through the model
We have classroom random effects for the intercept and slope
This means the prediction for an individual is made up of:
- Overall intercept
+ - Overall slope
+ - Classroom intercept offset (diff of classroom intercept from overall intercept)
+ - Classroom slope offset (diff of classroom slope from overall slope)
- Overall intercept
Next extract the ran_vals for the coresponding class
m2_ranvals <- tidy(m2, effects = "ran_vals")class1_ranvals <- m2_ranvals %>% filter(group == "class" & level == 1)class1_ranvals## # A tibble: 2 x 6## effect group level term estimate std.error## <chr> <chr> <chr> <chr> <dbl> <dbl>## 1 ran_vals class 1 (Intercept) 0.02657986 0.2240683## 2 ran_vals class 1 sexgirl -0.04272575 0.1956483Confirm with predict()
test <- popular %>% mutate(pred = predict(m2)) %>% filter(class == 10 & sex == "boy") test## # A tibble: 12 x 8## pupil class extrav sex texp popular popteach pred## <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>## 1 1 10 5 boy 21 5.4 5 4.675822## 2 3 10 5 boy 21 3.4 3 4.675822## 3 5 10 5 boy 21 5.1 7 4.675822## 4 7 10 5 boy 21 5.4 4 4.675822## 5 8 10 5 boy 21 4.9 4 4.675822## 6 11 10 6 boy 21 4.4 6 4.675822## 7 12 10 6 boy 21 4.9 5 4.675822## 8 14 10 6 boy 21 4.8 3 4.675822## 9 15 10 3 boy 21 3.6 3 4.675822## 10 16 10 5 boy 21 5 6 4.675822## 11 17 10 3 boy 21 5.5 5 4.675822## 12 18 10 6 boy 21 5 4 4.675822More on the predict function
- Once you have the model parameters, you can predict for any values of those parameters
Let's fit a slightly more complicated model, using a different longitudinal file than what you used (or are using) in the homework
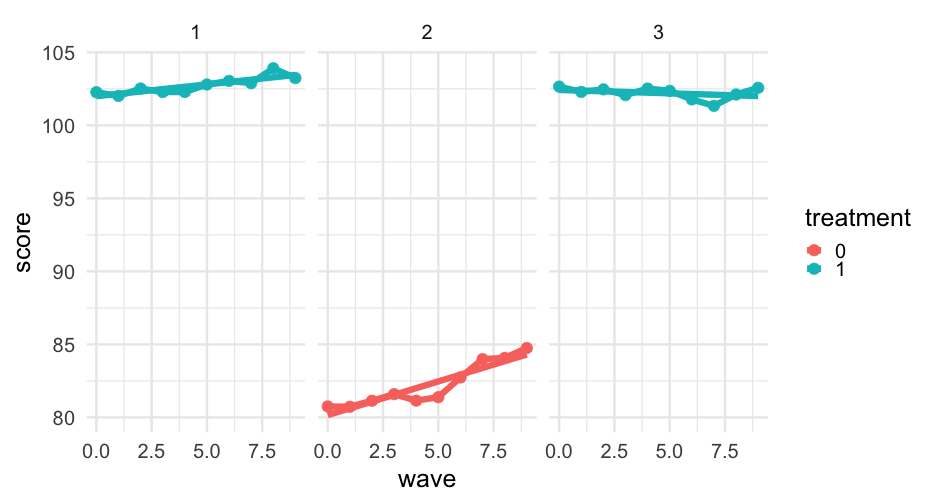
library(equatiomatic)head(sim_longitudinal)## # A tibble: 6 x 8## # Groups: school [1]## sid school district group treatment prop_low wave score## <int> <int> <int> <chr> <fct> <dbl> <dbl> <dbl>## 1 1 1 1 medium 1 0.1428571 0 102.2686## 2 1 1 1 medium 1 0.1428571 1 102.0135## 3 1 1 1 medium 1 0.1428571 2 102.5216## 4 1 1 1 medium 1 0.1428571 3 102.2792## 5 1 1 1 medium 1 0.1428571 4 102.2834## 6 1 1 1 medium 1 0.1428571 5 102.7963Plot predictions
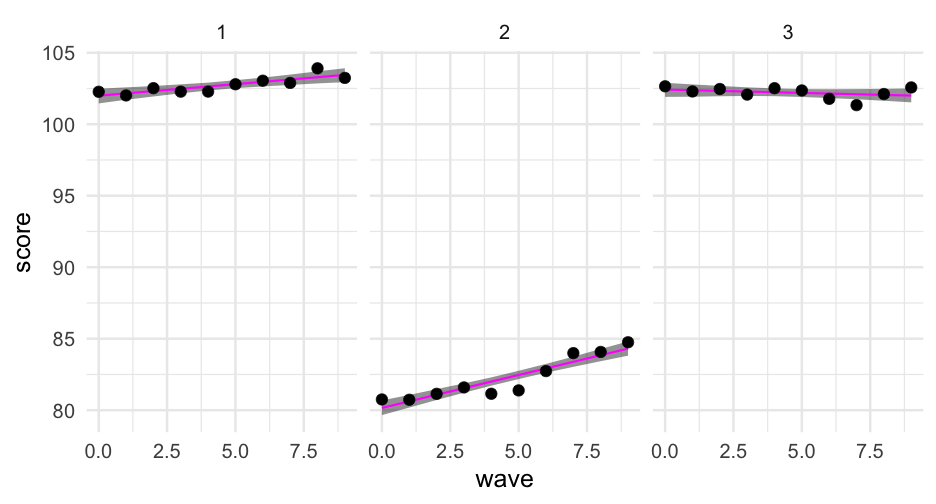
Let's first just limit our data to the first three students.
first_three <- sim_longitudinal %>% ungroup() %>% filter(sid %in% 1:3)Now try creating a new column in the data with the model predictions for these students. Specify newdata = first_three to only make the predictions for those cases.
02:00
first_three %>% mutate(model_pred = predict(m, newdata = first_three))## # A tibble: 30 x 9## sid school district group treatment prop_low wave score model_pred## <int> <int> <int> <chr> <fct> <dbl> <dbl> <dbl> <dbl>## 1 1 1 1 medium 1 0.1428571 0 102.2686 101.9955## 2 1 1 1 medium 1 0.1428571 1 102.0135 102.1572## 3 1 1 1 medium 1 0.1428571 2 102.5216 102.3189## 4 1 1 1 medium 1 0.1428571 3 102.2792 102.4806## 5 1 1 1 medium 1 0.1428571 4 102.2834 102.6423## 6 1 1 1 medium 1 0.1428571 5 102.7963 102.8040## 7 1 1 1 medium 1 0.1428571 6 103.0441 102.9657## 8 1 1 1 medium 1 0.1428571 7 102.8868 103.1274## 9 1 1 1 medium 1 0.1428571 8 103.9101 103.2891## 10 1 1 1 medium 1 0.1428571 9 103.2392 103.4508## # … with 20 more rowsCompare
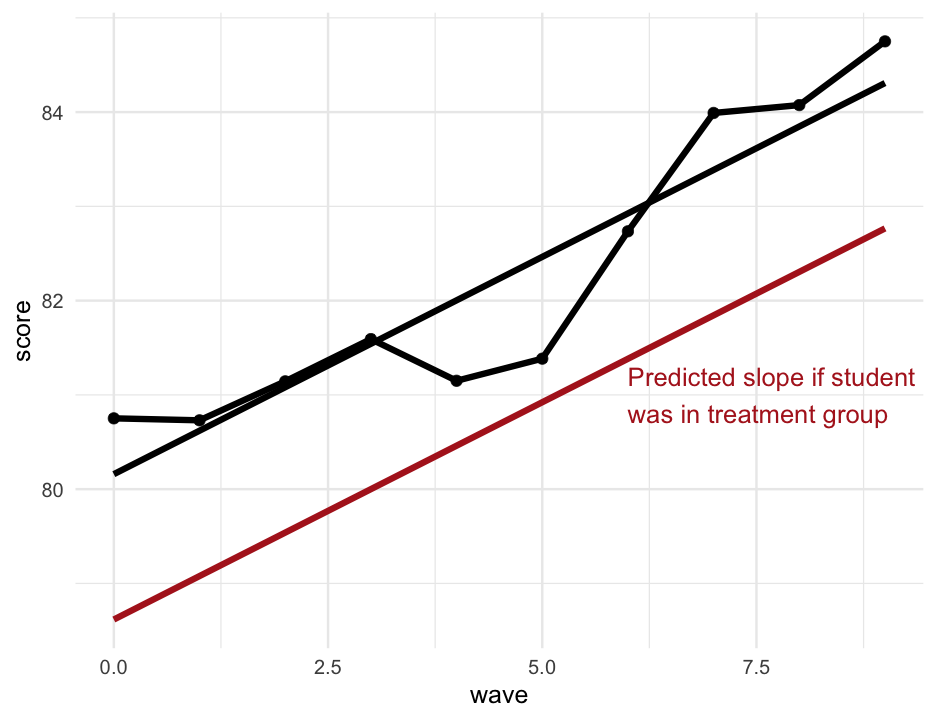
sim_longitudinal %>% filter(sid == 2) %>% mutate(model_pred = predict(m, newdata = .), trt_pred = predict(m, newdata = stu2_trt)) %>% ggplot(aes(wave, score)) + geom_point() + geom_line() + geom_line(aes(y = model_pred)) + geom_line(aes(y = trt_pred), color = "firebrick") + annotate( "text", x = 6, y = 81, hjust = 0, color = "firebrick", label = "Predicted slope if student\nwas in treatment group" )Wait... negative?
Yes...
arm::display(m)## lmer(formula = score ~ wave + treatment + (wave | sid), data = sim_longitudinal)## coef.est coef.se## (Intercept) 97.95 1.38 ## wave 0.17 0.03 ## treatment1 -1.54 1.92 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 9.74 ## wave 0.29 -0.16 ## Residual 0.44 ## ---## number of obs: 1000, groups: sid, 100## AIC = 2407.7, DIC = 2393.1## deviance = 2393.4Note - I'm just using an alternative function here to get it to fit on the slides easier
What to predict?
Let's say we want to predict what time points 10, 11, and 12 would look like for the first three students.
First, create the prediction data frame
pred_frame <- data.frame( sid = rep(1:3, each = 13), wave = rep(0:12), treatment = factor(rep(c(1, 0, 1), each = 13)))head(pred_frame)## sid wave treatment## 1 1 0 1## 2 1 1 1## 3 1 2 1## 4 1 3 1## 5 1 4 1## 6 1 5 1merTools
Next, load the {merTools} package
library(merTools)Create a prediction interval with predictInterval(), using simulation to obtain the prediction interval
m_pred_interval <- predictInterval( m, newdata = pred_frame, level = 0.95)m_pred_interval## fit upr lwr## 1 102.01207 104.72567 99.12815## 2 102.21023 104.90541 99.27463## 3 102.29546 105.12290 99.46212## 4 102.51889 105.24485 99.46114## 5 102.65108 105.36337 99.80513## 6 102.76418 105.52242 99.74719## 7 102.96957 105.60124 99.88279## 8 103.12596 105.79933 100.20903## 9 103.29646 105.97636 100.38468## 10 103.45665 106.11203 100.42733## 11 103.67235 106.41096 100.59995## 12 103.79786 106.50498 100.71008## 13 103.94205 106.78566 100.93741## 14 80.09841 82.96316 77.35830## 15 80.57764 83.33608 77.88836## 16 80.95438 83.81905 78.29942## 17 81.52330 84.29985 78.74763## 18 82.00058 84.70749 79.31407## 19 82.41634 85.17683 79.85975## 20 82.90433 85.69380 80.15920## 21 83.32670 86.17170 80.72806## 22 83.74488 86.63404 81.06728## 23 84.31655 87.24745 81.60025## 24 84.71815 87.79115 82.10399## 25 85.14727 88.28133 82.40308## 26 85.65959 88.65188 82.85234## 27 102.45376 105.27047 99.71076## 28 102.38756 105.17645 99.56086## 29 102.32892 105.19029 99.46407## 30 102.32501 104.91057 99.37041## 31 102.21161 104.91994 99.54185## 32 102.20953 104.91597 99.29393## 33 102.14807 104.84285 99.16836## 34 102.02224 104.84638 99.18406## 35 102.03473 104.67471 99.02696## 36 101.97196 104.73033 99.07564## 37 101.96588 104.69053 98.99436## 38 101.84893 104.56803 98.71735## 39 101.82603 104.67284 98.61854Binding data together
Let's add these predictions back to our prediction data frame, then plot them
bind_cols(pred_frame, m_pred_interval)## sid wave treatment fit upr lwr## 1 1 0 1 102.01207 104.72567 99.12815## 2 1 1 1 102.21023 104.90541 99.27463## 3 1 2 1 102.29546 105.12290 99.46212## 4 1 3 1 102.51889 105.24485 99.46114## 5 1 4 1 102.65108 105.36337 99.80513## 6 1 5 1 102.76418 105.52242 99.74719## 7 1 6 1 102.96957 105.60124 99.88279## 8 1 7 1 103.12596 105.79933 100.20903## 9 1 8 1 103.29646 105.97636 100.38468## 10 1 9 1 103.45665 106.11203 100.42733## 11 1 10 1 103.67235 106.41096 100.59995## 12 1 11 1 103.79786 106.50498 100.71008## 13 1 12 1 103.94205 106.78566 100.93741## 14 2 0 0 80.09841 82.96316 77.35830## 15 2 1 0 80.57764 83.33608 77.88836## 16 2 2 0 80.95438 83.81905 78.29942## 17 2 3 0 81.52330 84.29985 78.74763## 18 2 4 0 82.00058 84.70749 79.31407## 19 2 5 0 82.41634 85.17683 79.85975## 20 2 6 0 82.90433 85.69380 80.15920## 21 2 7 0 83.32670 86.17170 80.72806## 22 2 8 0 83.74488 86.63404 81.06728## 23 2 9 0 84.31655 87.24745 81.60025## 24 2 10 0 84.71815 87.79115 82.10399## 25 2 11 0 85.14727 88.28133 82.40308## 26 2 12 0 85.65959 88.65188 82.85234## 27 3 0 1 102.45376 105.27047 99.71076## 28 3 1 1 102.38756 105.17645 99.56086## 29 3 2 1 102.32892 105.19029 99.46407## 30 3 3 1 102.32501 104.91057 99.37041## 31 3 4 1 102.21161 104.91994 99.54185## 32 3 5 1 102.20953 104.91597 99.29393## 33 3 6 1 102.14807 104.84285 99.16836## 34 3 7 1 102.02224 104.84638 99.18406## 35 3 8 1 102.03473 104.67471 99.02696## 36 3 9 1 101.97196 104.73033 99.07564## 37 3 10 1 101.96588 104.69053 98.99436## 38 3 11 1 101.84893 104.56803 98.71735## 39 3 12 1 101.82603 104.67284 98.61854How?
What's really going on here?
See here for a full description, but basically:
- Creates a simulated (with
n.simsamples) distribution for each model parameter - Computes a distribution of predictions
- Returns the specifics of the prediction distribution, as requested (e.g.,
fitis the mean or median, anduprandlwrare the corresponding quaniteles for the uncertainty range)
Disclaimer
According to the {lme4} authors
There is no option for computing standard errors of predictions because it is difficult to define an efficient method that incorporates uncertainty in the variance parameters; we recommend lme4::bootMer() for this task.
The predictInterval() function is an approximation, while bootMer() is the "gold standard" (it just takes a long time)
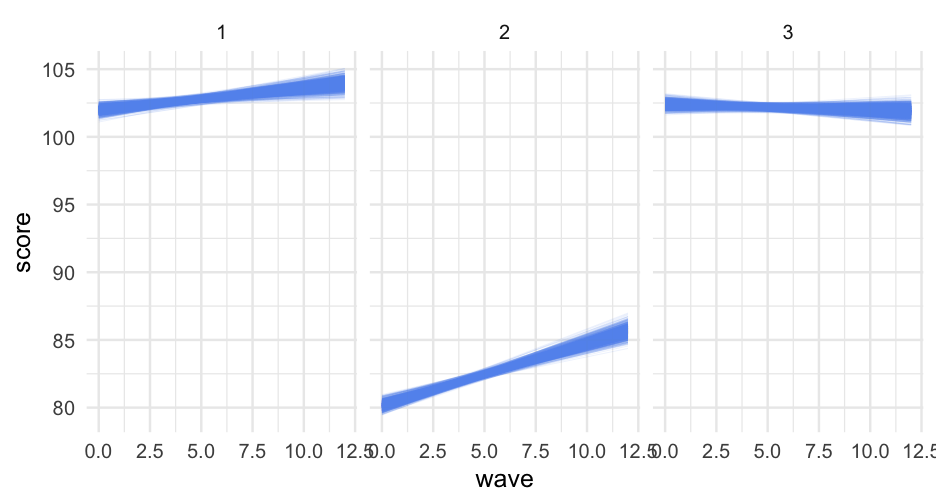
Now create BS Estimates
Note this takes a few seconds, and for bigger models may take a long time
b <- bootMer( m, nsim = 1000, FUN = pred_fun, use.u = TRUE, seed = 42)The predictions are stored in a matrix t
dim(b$t)## [1] 1000 39Each row of the matrix is the prediction for the given bootstrap estimate
Lots of options
There are lots of things you could do with this now. I'll move it to a data frame first
bd <- as.data.frame(t(b$t)) %>% mutate(sid = rep(1:3, each = 13), wave = rep(0:12, 3)) %>% pivot_longer( starts_with("V"), names_to = "bootstrap_sample", names_prefix = "V", names_transform = list(bootstrap_sample = as.numeric), values_to = "score" ) %>% arrange(sid, bootstrap_sample, wave)bd## # A tibble: 39,000 x 4## sid wave bootstrap_sample score## <int> <int> <dbl> <dbl>## 1 1 0 1 102.1749## 2 1 1 1 102.3497## 3 1 2 1 102.5245## 4 1 3 1 102.6992## 5 1 4 1 102.8740## 6 1 5 1 103.0488## 7 1 6 1 103.2235## 8 1 7 1 103.3983## 9 1 8 1 103.5731## 10 1 9 1 103.7478## # … with 38,990 more rowsPrefer ribbons?
bd_ribbons <- bd %>% group_by(sid, wave) %>% summarize(quantile = quantile(score, c(0.025, 0.975)), group = c("lower", "upper")) %>% pivot_wider(names_from = "group", values_from = "quantile")bd_ribbons## # A tibble: 39 x 4## # Groups: sid, wave [39]## sid wave lower upper## <int> <int> <dbl> <dbl>## 1 1 0 101.4553 102.4854## 2 1 1 101.7114 102.5782## 3 1 2 101.9408 102.6748## 4 1 3 102.1633 102.7898## 5 1 4 102.3576 102.9110## 6 1 5 102.5236 103.0809## 7 1 6 102.6428 103.2611## 8 1 7 102.7468 103.4728## 9 1 8 102.8555 103.6996## 10 1 9 102.9428 103.9153## # … with 29 more rowsImportant
There are different types of bootstrap resampling you can choose from
Do you want to randomly sample from the random effects also?
use.u = FALSEDo you want to assume the random effect estimates are true, and just sample the rest?
use.u = TRUE
Honestly - this is a bit confusing to me, and the results from use.u = FALSE didn't make much sense to me. But see here and here for more information
Adding additional levels
There are a few ways to do this:
- Assume your IDs are all unique
- implicit nesting
- Don't worry and the IDs, and specify the nesting through the formula
- Will result in equivalent estimates if the IDs are unique
See here for more examples
Implicit nesting
m1a <- lmer(score ~ wave*treatment + (wave|sid) + (1|school), data = sim_longitudinal)arm::display(m1a)## lmer(formula = score ~ wave * treatment + (wave | sid) + (1 | ## school), data = sim_longitudinal)## coef.est coef.se## (Intercept) 96.80 1.57 ## wave 0.40 0.03 ## treatment1 0.61 1.88 ## wave:treatment1 -0.45 0.04 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 9.16 ## wave 0.19 -0.13 ## school (Intercept) 3.27 ## Residual 0.44 ## ---## number of obs: 1000, groups: sid, 100; school, 15## AIC = 2330, DIC = 2301.2## deviance = 2306.6Explicit nesting
m1b <- lmer(score ~ wave*treatment + (wave|sid:school) + (1|school) , data = sim_longitudinal)arm::display(m1b)## lmer(formula = score ~ wave * treatment + (wave | sid:school) + ## (1 | school), data = sim_longitudinal)## coef.est coef.se## (Intercept) 96.80 1.57 ## wave 0.40 0.03 ## treatment1 0.61 1.88 ## wave:treatment1 -0.45 0.04 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid:school (Intercept) 9.16 ## wave 0.19 -0.13 ## school (Intercept) 3.27 ## Residual 0.44 ## ---## number of obs: 1000, groups: sid:school, 100; school, 15## AIC = 2330, DIC = 2301.2## deviance = 2306.6Predictions "by hand"
This is now a three-level model. How does it differ from two-level models in terms of model predictions?
Let's make a prediction for the first student at the fourth time point:
sim_longitudinal[4, ]## # A tibble: 1 x 8## # Groups: school [1]## sid school district group treatment prop_low wave score## <int> <int> <int> <chr> <fct> <dbl> <dbl> <dbl>## 1 1 1 1 medium 1 0.1428571 3 102.2792The pieces
fixed <- fixef(m1a)ranefs <- ranef(m1a)fixed## (Intercept) wave treatment1 wave:treatment1 ## 96.7973387 0.4010222 0.6080336 -0.4459053# Pull just the ranefs for sid 1 and school 1sid_ranefs <- ranefs$sid[1, ] sid_ranefs## (Intercept) wave## 1 1.85279 0.1936603sch_ranefs <- ranefs$school[1, ]sch_ranefs## [1] 2.795928Predictions
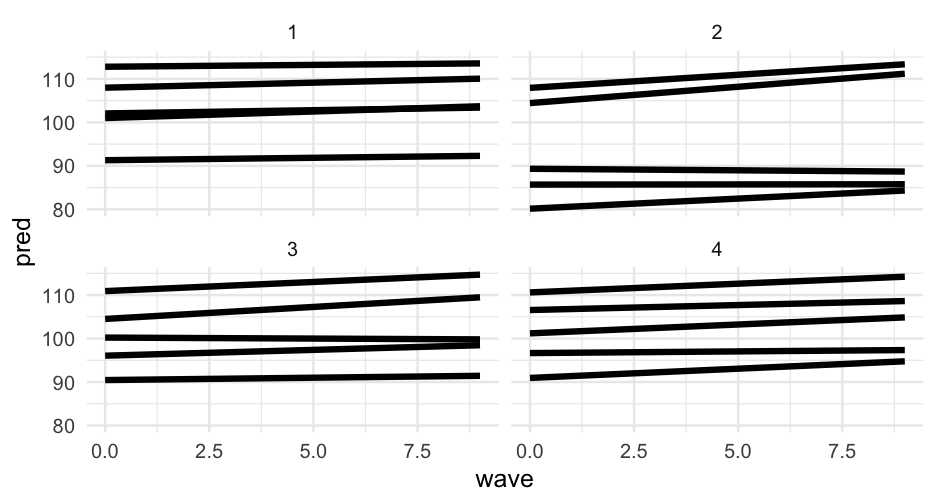
Let's randomly sample 5 students in the first four school and display the model predictions for those students.
There are many ways to create the sample, here's one:
samp <- sim_longitudinal %>% filter(school %in% 1:4) %>% group_by(school, sid) %>% nest()samp## # A tibble: 28 x 3## # Groups: sid, school [28]## sid school data ## <int> <int> <list> ## 1 1 1 <tibble [10 × 6]>## 2 33 3 <tibble [10 × 6]>## 3 34 4 <tibble [10 × 6]>## 4 46 1 <tibble [10 × 6]>## 5 61 1 <tibble [10 × 6]>## 6 77 2 <tibble [10 × 6]>## 7 78 3 <tibble [10 × 6]>## 8 91 1 <tibble [10 × 6]>## 9 93 3 <tibble [10 × 6]>## 10 2 2 <tibble [10 × 6]>## # … with 18 more rowsSelect 5 rows for each school
set.seed(42)samp %>% group_by(school) %>% sample_n(5)## # A tibble: 20 x 3## # Groups: school [4]## sid school data ## <int> <int> <list> ## 1 1 1 <tibble [10 × 6]>## 2 76 1 <tibble [10 × 6]>## 3 31 1 <tibble [10 × 6]>## 4 16 1 <tibble [10 × 6]>## 5 46 1 <tibble [10 × 6]>## 6 62 2 <tibble [10 × 6]>## 7 2 2 <tibble [10 × 6]>## 8 92 2 <tibble [10 × 6]>## 9 77 2 <tibble [10 × 6]>## 10 47 2 <tibble [10 × 6]>## 11 63 3 <tibble [10 × 6]>## 12 18 3 <tibble [10 × 6]>## 13 33 3 <tibble [10 × 6]>## 14 48 3 <tibble [10 × 6]>## 15 78 3 <tibble [10 × 6]>## 16 19 4 <tibble [10 × 6]>## 17 49 4 <tibble [10 × 6]>## 18 4 4 <tibble [10 × 6]>## 19 79 4 <tibble [10 × 6]>## 20 94 4 <tibble [10 × 6]>unnest()
set.seed(42)samp <- samp %>% group_by(school) %>% sample_n(5) %>% unnest(data) %>% ungroup()samp## # A tibble: 200 x 8## sid school district group treatment prop_low wave score## <int> <int> <int> <chr> <fct> <dbl> <dbl> <dbl>## 1 1 1 1 medium 1 0.1428571 0 102.2686## 2 1 1 1 medium 1 0.1428571 1 102.0135## 3 1 1 1 medium 1 0.1428571 2 102.5216## 4 1 1 1 medium 1 0.1428571 3 102.2792## 5 1 1 1 medium 1 0.1428571 4 102.2834## 6 1 1 1 medium 1 0.1428571 5 102.7963## 7 1 1 1 medium 1 0.1428571 6 103.0441## 8 1 1 1 medium 1 0.1428571 7 102.8868## 9 1 1 1 medium 1 0.1428571 8 103.9101## 10 1 1 1 medium 1 0.1428571 9 103.2392## # … with 190 more rowsMake prediction
samp %>% mutate(pred = predict(m1a, newdata = samp))## # A tibble: 200 x 9## sid school district group treatment prop_low wave score pred## <int> <int> <int> <chr> <fct> <dbl> <dbl> <dbl> <dbl>## 1 1 1 1 medium 1 0.1428571 0 102.2686 102.0541## 2 1 1 1 medium 1 0.1428571 1 102.0135 102.2029## 3 1 1 1 medium 1 0.1428571 2 102.5216 102.3516## 4 1 1 1 medium 1 0.1428571 3 102.2792 102.5004## 5 1 1 1 medium 1 0.1428571 4 102.2834 102.6492## 6 1 1 1 medium 1 0.1428571 5 102.7963 102.7980## 7 1 1 1 medium 1 0.1428571 6 103.0441 102.9468## 8 1 1 1 medium 1 0.1428571 7 102.8868 103.0955## 9 1 1 1 medium 1 0.1428571 8 103.9101 103.2443## 10 1 1 1 medium 1 0.1428571 9 103.2392 103.3931## # … with 190 more rowsMarginal effect
A marginal effect shows the relation between one variable in the model and the outcome, while averaging over the other predictors in the model
Let's start by making the models slightly more complicated
m2 <- lmer(score ~ wave*treatment + group + prop_low + (wave|sid) + (1|school) , data = sim_longitudinal)arm::display(m2, detail = TRUE)## lmer(formula = score ~ wave * treatment + group + prop_low + ## (wave | sid) + (1 | school), data = sim_longitudinal)## coef.est coef.se t value## (Intercept) 97.62 4.70 20.78 ## wave 0.40 0.03 14.38 ## treatment1 0.82 1.91 0.43 ## grouplow -4.83 4.00 -1.21 ## groupmedium -3.61 3.75 -0.96 ## prop_low 9.34 10.21 0.91 ## wave:treatment1 -0.45 0.04 -11.42 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 9.18 ## wave 0.19 -0.13 ## school (Intercept) 3.41 ## Residual 0.44 ## ---## number of obs: 1000, groups: sid, 100; school, 15## AIC = 2319.8, DIC = 2313.2## deviance = 2304.5Marginal effect
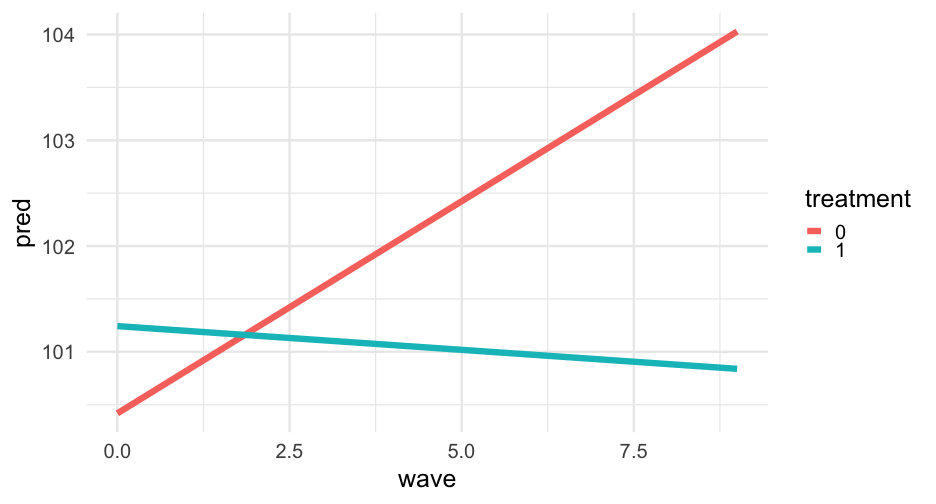
Let's look at the relation between wave and score by treatment, holding the other values constant.
First, build a prediction data frame. We'll make population-level prediction - i.e., ignoring the random effects.
marginal_frame1 <- data.frame( wave = rep(0:9, 2), treatment = as.factor(rep(c(0, 1), each = 10)), group = factor("high", levels = c("low", "medium", "high")), prop_low = mean(sim_longitudinal$prop_low, na.rm = TRUE), sid = -999, school = -999)Marginal effect
Let's look at the relation between wave and score by treatment, holding the other values constant.
First, build a prediction data frame. We'll make population-level prediction - i.e., ignoring the random effects.
marginal_frame1 <- data.frame( wave = rep(0:9, 2), treatment = as.factor(rep(c(0, 1), each = 10)), group = factor("high", levels = c("low", "medium", "high")), prop_low = mean(sim_longitudinal$prop_low, na.rm = TRUE), sid = -999, school = -999)The length of each of these variables can sometimes get a little tricky
marginal_frame1## wave treatment group prop_low sid school## 1 0 0 high 0.3 -999 -999## 2 1 0 high 0.3 -999 -999## 3 2 0 high 0.3 -999 -999## 4 3 0 high 0.3 -999 -999## 5 4 0 high 0.3 -999 -999## 6 5 0 high 0.3 -999 -999## 7 6 0 high 0.3 -999 -999## 8 7 0 high 0.3 -999 -999## 9 8 0 high 0.3 -999 -999## 10 9 0 high 0.3 -999 -999## 11 0 1 high 0.3 -999 -999## 12 1 1 high 0.3 -999 -999## 13 2 1 high 0.3 -999 -999## 14 3 1 high 0.3 -999 -999## 15 4 1 high 0.3 -999 -999## 16 5 1 high 0.3 -999 -999## 17 6 1 high 0.3 -999 -999## 18 7 1 high 0.3 -999 -999## 19 8 1 high 0.3 -999 -999## 20 9 1 high 0.3 -999 -999Make predictions!
Note that we have to specify to allow new levels
marginal_frame1 <- marginal_frame1 %>% mutate(pred = predict(m2, newdata = marginal_frame1, allow.new.levels = TRUE))marginal_frame1## wave treatment group prop_low sid school pred## 1 0 0 high 0.3 -999 -999 100.4186## 2 1 0 high 0.3 -999 -999 100.8196## 3 2 0 high 0.3 -999 -999 101.2207## 4 3 0 high 0.3 -999 -999 101.6217## 5 4 0 high 0.3 -999 -999 102.0227## 6 5 0 high 0.3 -999 -999 102.4237## 7 6 0 high 0.3 -999 -999 102.8248## 8 7 0 high 0.3 -999 -999 103.2258## 9 8 0 high 0.3 -999 -999 103.6268## 10 9 0 high 0.3 -999 -999 104.0278## 11 0 1 high 0.3 -999 -999 101.2433## 12 1 1 high 0.3 -999 -999 101.1984## 13 2 1 high 0.3 -999 -999 101.1536## 14 3 1 high 0.3 -999 -999 101.1087## 15 4 1 high 0.3 -999 -999 101.0638## 16 5 1 high 0.3 -999 -999 101.0189## 17 6 1 high 0.3 -999 -999 100.9740## 18 7 1 high 0.3 -999 -999 100.9291## 19 8 1 high 0.3 -999 -999 100.8843## 20 9 1 high 0.3 -999 -999 100.8394Try again
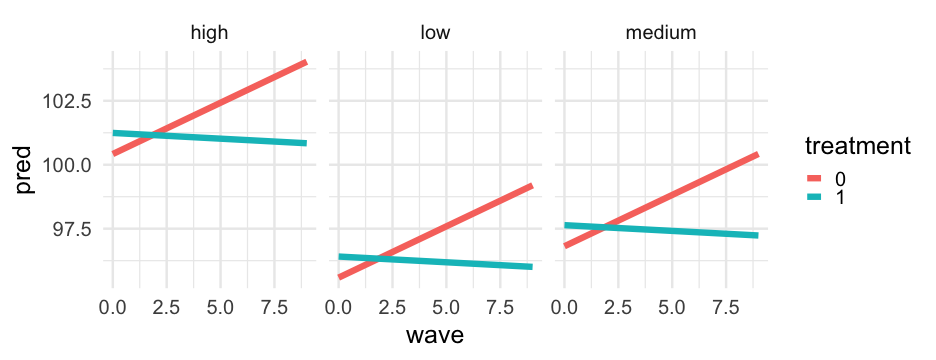
This time, let's look at the same thing, but for all groups, still holding prop_low constant.
marginal_frame2 <- data.frame( wave = rep(0:9, 2*3), treatment = as.factor(rep(c(0, 1), each = 10*3)), group = factor( rep( rep(c("low", "medium", "high"), each = 10), 2 ) ), prop_low = mean(sim_longitudinal$prop_low, na.rm = TRUE), sid = -999, school = -999)Automated method
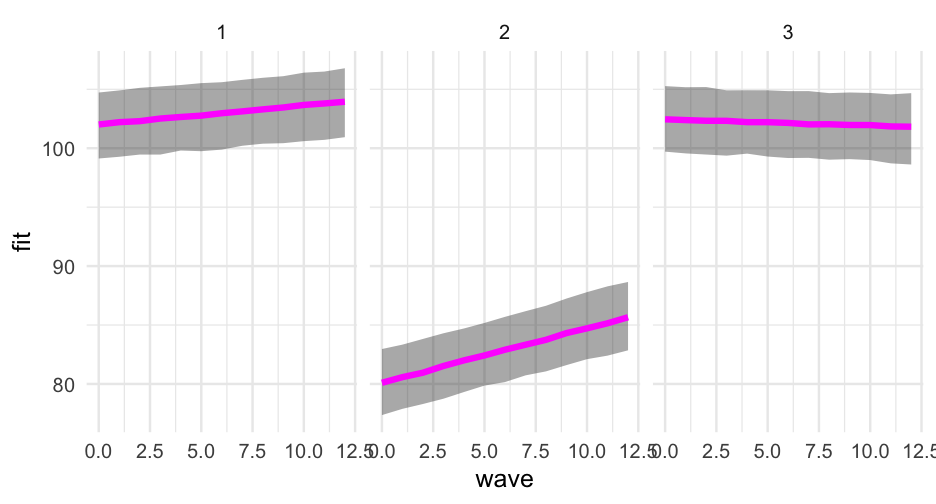
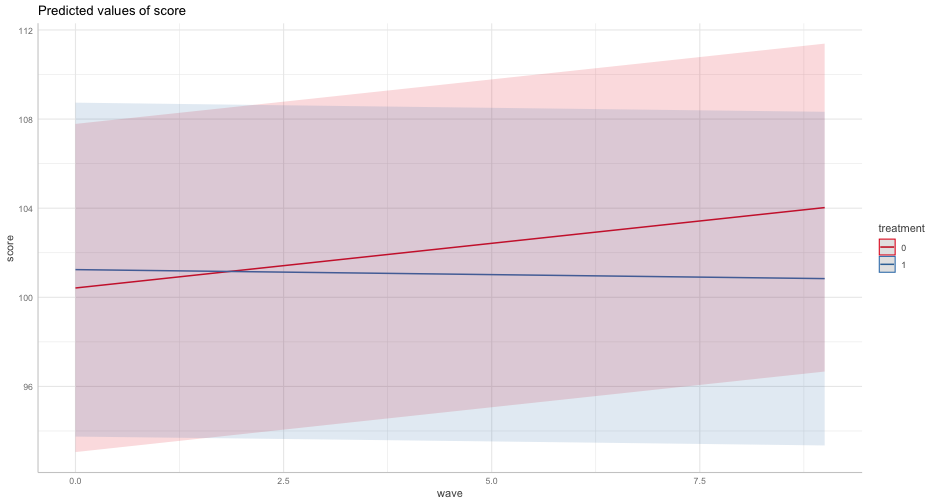
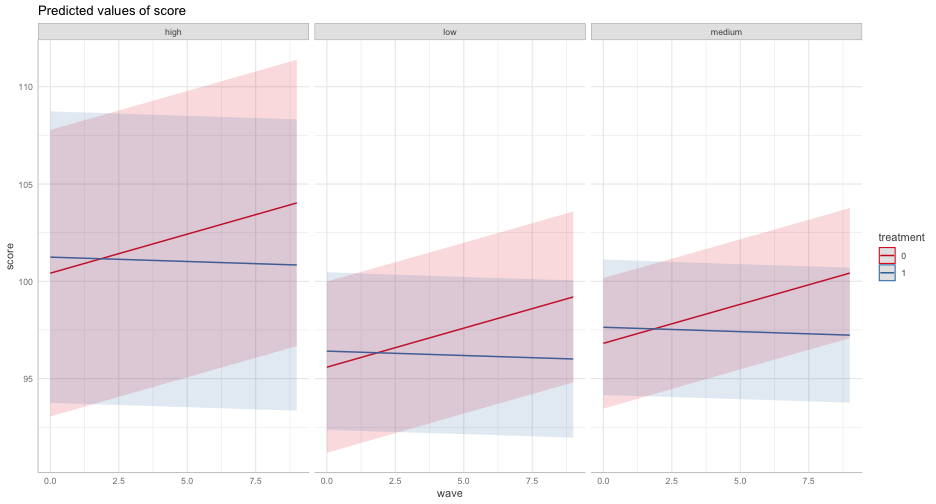
We can build this up on our own with new data frames
library(ggeffects)ggpredict(m2, "wave")## # Predicted values of score## # x = wave## ## x | Predicted | 95% CI## -------------------------------## 0 | 100.42 | [93.05, 107.78]## 1 | 100.82 | [93.46, 108.18]## 2 | 101.22 | [93.86, 108.58]## 3 | 101.62 | [94.26, 108.98]## 5 | 102.42 | [95.07, 109.78]## 6 | 102.82 | [95.47, 110.18]## 7 | 103.23 | [95.87, 110.58]## 9 | 104.03 | [96.67, 111.39]## ## Adjusted for:## * treatment = 0## * group = high## * prop_low = 0.30## * sid = 0 (population-level)## * school = 0 (population-level)Individual observation
We can get these prediction intervals for specific levels also
ggpredict(m2, "wave", condition = c(sid = 1, school = 1))## # Predicted values of score## # x = wave## ## x | Predicted | 95% CI## -------------------------------## 0 | 100.42 | [93.05, 107.78]## 1 | 100.82 | [93.46, 108.18]## 2 | 101.22 | [93.86, 108.58]## 3 | 101.62 | [94.26, 108.98]## 5 | 102.42 | [95.07, 109.78]## 6 | 102.82 | [95.47, 110.18]## 7 | 103.23 | [95.87, 110.58]## 9 | 104.03 | [96.67, 111.39]## ## Adjusted for:## * treatment = 0## * group = high## * prop_low = 0.30