Variance-Covariance Matrices
Daniel Anderson
Week 4
Learning Objectives
Gain a deeper understanding of how the residual structure is different in multilevel models
Understand that there are methods for changing the residual structure, and understand when and why this might be preferable
Be able to implement alternative methods using {nlme}
New data
sim3 <- read_csv(here::here("data", "sim3level.csv"))sim3## # A tibble: 570 x 6## Math ActiveTime ClassSize Classroom School StudentID## <dbl> <dbl> <dbl> <dbl> <chr> <dbl>## 1 55.42604 0.06913359 18 1 Sch1 1## 2 54.34306 0.08462063 18 1 Sch1 2## 3 61.42570 0.1299456 18 1 Sch1 3## 4 56.12271 0.7461320 18 1 Sch1 4## 5 53.34900 0.03887918 18 1 Sch1 5## 6 57.99773 0.6856354 18 1 Sch1 6## 7 54.46326 0.1439774 18 1 Sch1 7## 8 64.20517 0.8910800 18 1 Sch1 8## 9 54.49117 0.08963612 18 1 Sch1 9## 10 57.87599 0.03773272 18 1 Sch1 10## # … with 560 more rows01:00
Data obtained here
Make classroom unique
We could handle this with our model syntax, but nested IDs like this always makes me nervous. Let's make them unique.
sim3 <- sim3 %>% mutate(class_id = paste0("class", Classroom, ":", School))sim3## # A tibble: 570 x 7## Math ActiveTime ClassSize Classroom School StudentID class_id ## <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr> ## 1 55.42604 0.06913359 18 1 Sch1 1 class1:Sch1## 2 54.34306 0.08462063 18 1 Sch1 2 class1:Sch1## 3 61.42570 0.1299456 18 1 Sch1 3 class1:Sch1## 4 56.12271 0.7461320 18 1 Sch1 4 class1:Sch1## 5 53.34900 0.03887918 18 1 Sch1 5 class1:Sch1## 6 57.99773 0.6856354 18 1 Sch1 6 class1:Sch1## 7 54.46326 0.1439774 18 1 Sch1 7 class1:Sch1## 8 64.20517 0.8910800 18 1 Sch1 8 class1:Sch1## 9 54.49117 0.08963612 18 1 Sch1 9 class1:Sch1## 10 57.87599 0.03773272 18 1 Sch1 10 class1:Sch1## # … with 560 more rowsThis one
What about this one?
02:00
Mathi∼N(αj[i],k[i]+β1j[i],k[i](ActiveTime),σ2)(αjβ1j)∼N((γα0+γα1(ClassSize)μβ1j),(σ2αjραjβ1jρβ1jαjσ2β1j)), for class_id j = 1,…,J(αkβ1k)∼N((μαkμβ1k),(σ2αkραkβ1kρβ1kαkσ2β1k)), for School k = 1,…,K
lmer(Math ~ ActiveTime + ClassSize + (ActiveTime|class_id) + (ActiveTime|School), data = sim3)Last one
What about this one?
02:00
Mathi∼N(αj[i],k[i]+β1j[i],k[i](ActiveTime),σ2)(αjβ1j)∼N((γα0+γα1k[i](ClassSize)γβ10+γβ11(ClassSize)),(σ2αjραjβ1jρβ1jαjσ2β1j)), for class_id j = 1,…,J⎛⎜⎝αkβ1kγ1k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝μαkμβ1kμγ1k⎞⎟⎠,⎛⎜ ⎜⎝σ2αkραkβ1kραkγ1kρβ1kαkσ2β1kρβ1kγ1kργ1kαkργ1kβ1kσ2γ1k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for School k = 1,…,K
Last one
What about this one?
02:00
Mathi∼N(αj[i],k[i]+β1j[i],k[i](ActiveTime),σ2)(αjβ1j)∼N((γα0+γα1k[i](ClassSize)γβ10+γβ11(ClassSize)),(σ2αjραjβ1jρβ1jαjσ2β1j)), for class_id j = 1,…,J⎛⎜⎝αkβ1kγ1k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝μαkμβ1kμγ1k⎞⎟⎠,⎛⎜ ⎜⎝σ2αkραkβ1kραkγ1kρβ1kαkσ2β1kρβ1kγ1kργ1kαkργ1kβ1kσ2γ1k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for School k = 1,…,K
lmer(Math ~ ActiveTime * ClassSize + (ActiveTime|class_id) + (ActiveTime + ClassSize|School), data = sim3)Data
- n = 35 people
- Each completed a cognitive inventory on "opposites naming"
- At first time point, participants also completed a general cognitive measure
Read in data
willett <- read_csv(here::here("data", "willett-1988.csv"))willett## # A tibble: 140 x 4## id time opp cog## <dbl> <dbl> <dbl> <dbl>## 1 1 0 205 137## 2 1 1 217 137## 3 1 2 268 137## 4 1 3 302 137## 5 2 0 219 123## 6 2 1 243 123## 7 2 2 279 123## 8 2 3 302 123## 9 3 0 142 129## 10 3 1 212 129## # … with 130 more rowsStandard OLS
We have four observations per participant.
If we fit a standard OLS model, it would look like this
bad <- lm(opp ~ time, data = willett)summary(bad)## ## Call:## lm(formula = opp ~ time, data = willett)## ## Residuals:## Min 1Q Median 3Q Max ## -88.374 -25.584 1.186 28.926 64.746 ## ## Coefficients:## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 164.374 5.035 32.65 <2e-16 ***## time 26.960 2.691 10.02 <2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## ## Residual standard error: 35.6 on 138 degrees of freedom## Multiple R-squared: 0.421, Adjusted R-squared: 0.4168 ## F-statistic: 100.3 on 1 and 138 DF, p-value: < 2.2e-16Individual level residuals
We can expand our notation, so it looks like a multivariate normal distribution
⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝ϵ1 ϵ2 ϵ3 ⋮ ϵn⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠∼MVN⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣0 0 0 ⋮ 0⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦,⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣σϵ00…0 0σϵ000 00σϵ00 ⋮00⋱⋮ 000…σϵ⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠
This is where the i.i.d. part comes in. The residuals are assumed independent and identically distributed.
Multilevel model
Very regularly, there are reasons to believe the i.i.d. assumption is violated. Consider our current case, with 4 time points for each individual.
Is an observation for one time point for one individual independent from the other observations for that individual?
Rather than estimating a single residual variance, we estimate an additional components associated with individuals, leading to a block diagonal structure
Block diagonal
⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝ϵ11ϵ12ϵ13ϵ14ϵ21ϵ22ϵ23ϵ24⋮ϵn1ϵn2ϵn3ϵn4⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠∼⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣00000000⋮0000⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦,⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣σ11σ12σ13σ140000…0000σ21σ22σ23σ240000…0000σ31σ32σ33σ340000…0000σ41σ42σ43σ440000…00000000σ11σ12σ13σ14…00000000σ21σ22σ23σ24…00000000σ31σ32σ33σ34…00000000σ41σ42σ43σ44…0000⋮⋮⋮⋮⋮⋮⋮⋮⋱⋮⋮⋮⋮00000000…σ11σ12σ13σ1400000000…σ21σ22σ23σ2400000000…σ31σ32σ33σ3400000000…σ41σ42σ43σ44⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠
Block diagonal
⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝ϵ11ϵ12ϵ13ϵ14ϵ21ϵ22ϵ23ϵ24⋮ϵn1ϵn2ϵn3ϵn4⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠∼⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣00000000⋮0000⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦,⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣σ11σ12σ13σ140000…0000σ21σ22σ23σ240000…0000σ31σ32σ33σ340000…0000σ41σ42σ43σ440000…00000000σ11σ12σ13σ14…00000000σ21σ22σ23σ24…00000000σ31σ32σ33σ34…00000000σ41σ42σ43σ44…0000⋮⋮⋮⋮⋮⋮⋮⋮⋱⋮⋮⋮⋮00000000…σ11σ12σ13σ1400000000…σ21σ22σ23σ2400000000…σ31σ32σ33σ3400000000…σ41σ42σ43σ44⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠
Correlations for off-diagonals estimated
Block diagonal
⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝ϵ11ϵ12ϵ13ϵ14ϵ21ϵ22ϵ23ϵ24⋮ϵn1ϵn2ϵn3ϵn4⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠∼⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣00000000⋮0000⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦,⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣σ11σ12σ13σ140000…0000σ21σ22σ23σ240000…0000σ31σ32σ33σ340000…0000σ41σ42σ43σ440000…00000000σ11σ12σ13σ14…00000000σ21σ22σ23σ24…00000000σ31σ32σ33σ34…00000000σ41σ42σ43σ44…0000⋮⋮⋮⋮⋮⋮⋮⋮⋱⋮⋮⋮⋮00000000…σ11σ12σ13σ1400000000…σ21σ22σ23σ2400000000…σ31σ32σ33σ3400000000…σ41σ42σ43σ44⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠
Correlations for off-diagonals estimated
Same variance components for all blocks
Block diagonal
⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝ϵ11ϵ12ϵ13ϵ14ϵ21ϵ22ϵ23ϵ24⋮ϵn1ϵn2ϵn3ϵn4⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠∼⎛⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜ ⎜⎝⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣00000000⋮0000⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦,⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣σ11σ12σ13σ140000…0000σ21σ22σ23σ240000…0000σ31σ32σ33σ340000…0000σ41σ42σ43σ440000…00000000σ11σ12σ13σ14…00000000σ21σ22σ23σ24…00000000σ31σ32σ33σ34…00000000σ41σ42σ43σ44…0000⋮⋮⋮⋮⋮⋮⋮⋮⋱⋮⋮⋮⋮00000000…σ11σ12σ13σ1400000000…σ21σ22σ23σ2400000000…σ31σ32σ33σ3400000000…σ41σ42σ43σ44⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎞⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟ ⎟⎠
Correlations for off-diagonals estimated
Same variance components for all blocks
Out-of-block diagonals are still zero
Homogeneity of variance
As mentioned on the previous slide, we assume the same variance components across all student
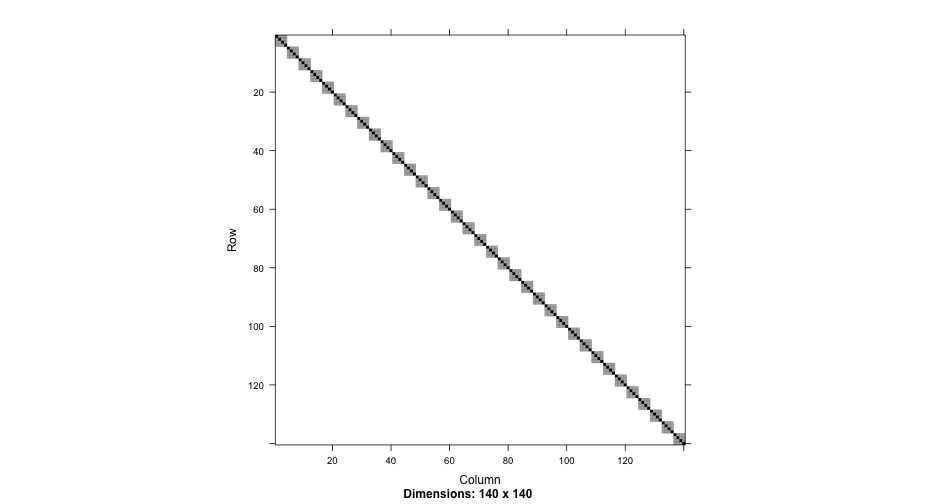
This is referred to as the homogeneity of variance assumption - although the block (often referred to as the composite residual) may be heteroscedastic and dependent within a grouping factor (i.e., people) the entire error structure is repeated identically across units (i.e., people)
Pull first few rows/cols
w0_rvcv[1:8, 1:8]## 8 x 8 sparse Matrix of class "dgCMatrix"## 1 2 3 4 5 6 7 8## 1 1280.7065 904.8054 904.8054 904.8054 . . . . ## 2 904.8054 1280.7065 904.8054 904.8054 . . . . ## 3 904.8054 904.8054 1280.7065 904.8054 . . . . ## 4 904.8054 904.8054 904.8054 1280.7065 . . . . ## 5 . . . . 1280.7065 904.8054 904.8054 904.8054## 6 . . . . 904.8054 1280.7065 904.8054 904.8054## 7 . . . . 904.8054 904.8054 1280.7065 904.8054## 8 . . . . 904.8054 904.8054 904.8054 1280.7065Understanding these numbers
Let's look at the model output
arm::display(w0)## lmer(formula = opp ~ time + (1 | id), data = willett)## coef.est coef.se## (Intercept) 164.37 5.78 ## time 26.96 1.47 ## ## Error terms:## Groups Name Std.Dev.## id (Intercept) 30.08 ## Residual 19.39 ## ---## number of obs: 140, groups: id, 35## AIC = 1308.3, DIC = 1315.9## deviance = 1308.1The diagonal values were 1280.7065389 while the off diagonal values were 904.8053852
Let's extract the variance components from our model.
vars_w0 <- as.data.frame(VarCorr(w0))vars_w0## grp var1 var2 vcov sdcor## 1 id (Intercept) <NA> 904.8054 30.07998## 2 Residual <NA> <NA> 375.9012 19.38817The diagonal values were 1280.7065389 while the off diagonal values were 904.8053852
Let's extract the variance components from our model.
vars_w0 <- as.data.frame(VarCorr(w0))vars_w0## grp var1 var2 vcov sdcor## 1 id (Intercept) <NA> 904.8054 30.07998## 2 Residual <NA> <NA> 375.9012 19.38817Notice anything?
The diagonal values were 1280.7065389 while the off diagonal values were 904.8053852
Let's extract the variance components from our model.
vars_w0 <- as.data.frame(VarCorr(w0))vars_w0## grp var1 var2 vcov sdcor## 1 id (Intercept) <NA> 904.8054 30.07998## 2 Residual <NA> <NA> 375.9012 19.38817Notice anything?
The diagonals are given by sum(vars_w0)$vcov while the off-diagonals are just the intercept variance
The composite residual
w1 <- lmer(opp ~ time + (time|id), willett)w1_rvcv <- pull_residual_vcov(w1)w1_rvcv[1:4, 1:4]## 4 x 4 sparse Matrix of class "dgCMatrix"## 1 2 3 4## 1 1358.2469 1019.5095 840.2515 660.9934## 2 1019.5095 1132.1294 925.7905 878.9310## 3 840.2515 925.7905 1170.8089 1096.8686## 4 660.9934 878.9310 1096.8686 1474.2856The composite residual
w1 <- lmer(opp ~ time + (time|id), willett)w1_rvcv <- pull_residual_vcov(w1)w1_rvcv[1:4, 1:4]## 4 x 4 sparse Matrix of class "dgCMatrix"## 1 2 3 4## 1 1358.2469 1019.5095 840.2515 660.9934## 2 1019.5095 1132.1294 925.7905 878.9310## 3 840.2515 925.7905 1170.8089 1096.8686## 4 660.9934 878.9310 1096.8686 1474.2856Unstructured
The model we fit has an unstructured variance co-variance matrix. While each block is the same, every element of the block is now estimated.
Calculate
diag(w1_rvcv[1:4, 1:4])## [1] 1358.247 1132.129 1170.809 1474.286residual + int_var## [1] 1358.247residual + int_var + 2*covar + slope_var## [1] 1132.129residual + int_var + (2*covar)*2 + slope_var*2^2## [1] 1170.809residual + int_var + (2*covar)*3 + slope_var*3^2## [1] 1474.286Calculate a few
w1_rvcv[1:4, 1:4]## 4 x 4 sparse Matrix of class "dgCMatrix"## 1 2 3 4## 1 1358.2469 1019.5095 840.2515 660.9934## 2 1019.5095 1132.1294 925.7905 878.9310## 3 840.2515 925.7905 1170.8089 1096.8686## 4 660.9934 878.9310 1096.8686 1474.2856int_var + covar*(1 + 0) + slope_var*1*0## [1] 1019.51int_var + covar*(2 + 1) + slope_var*2*1## [1] 925.7905int_var + covar*(3 + 2) + slope_var*3*2## [1] 1096.869int_var + covar*(2 + 0) + slope_var*2*0## [1] 840.2515The possibilities
There are a number of alternative structures. We'll talk about a few here.
If you want to go deeper, I suggest Singer & Willett, Chapter 7
Code to fit models with each type of structure, using the same Willett data we're using today, is available here
Structures we'll fit
Unstructured (default with lme4, we've already seen this)
Variance components (no off-diagonals)
Autoregressive
Heterogeneous autoregressive
Toeplitz
Note this is just a sampling. Other structures are possible.
Outside of unstructured & variance component only models, we'll need to use the nlme package
Structures we'll fit
Unstructured (default with lme4, we've already seen this)
Variance components (no off-diagonals)
Autoregressive
Heterogeneous autoregressive
Toeplitz
Note this is just a sampling. Other structures are possible.
Outside of unstructured & variance component only models, we'll need to use the nlme package
We could also use Bayes, as we'll get to in a few weeks.
The model
Generally, in a model like the Willett data, we would estimate intercept variance, and maybe slope variance.
We saw earlier how these combine to create the residual variance-covariance matrix
Alternatively, we can estimate separate variances at each time point
Σr=⎡⎢ ⎢ ⎢ ⎢ ⎢⎣σ210000σ220000σ230000σ24⎤⎥ ⎥ ⎥ ⎥ ⎥⎦
One-hot encoding
First, use one-hot encoding for time
Same as dummy-coding, but you use all the levels.
w <- willett %>% mutate(t0 = ifelse(time == 0, 1, 0), t1 = ifelse(time == 1, 1, 0), t2 = ifelse(time == 2, 1, 0), t3 = ifelse(time == 3, 1, 0))w## # A tibble: 140 x 8## id time opp cog t0 t1 t2 t3## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 0 205 137 1 0 0 0## 2 1 1 217 137 0 1 0 0## 3 1 2 268 137 0 0 1 0## 4 1 3 302 137 0 0 0 1## 5 2 0 219 123 1 0 0 0## 6 2 1 243 123 0 1 0 0## 7 2 2 279 123 0 0 1 0## 8 2 3 302 123 0 0 0 1## 9 3 0 142 129 1 0 0 0## 10 3 1 212 129 0 1 0 0## # … with 130 more rowsAlternative creation
The model.matrix() function automatically dummy-codes factors.
model.matrix( ~ 0 + factor(time), data = willett) %>% head()## factor(time)0 factor(time)1 factor(time)2 factor(time)3## 1 1 0 0 0## 2 0 1 0 0## 3 0 0 1 0## 4 0 0 0 1## 5 1 0 0 0## 6 0 1 0 0Could be helpful if time is coded in a more complicated way
Fit the model
varcomp <- lmer(opp ~ time + (0 + t0 + t1 + t2 + t3 || id), w)summary(varcomp)## Linear mixed model fit by REML ['lmerMod']## Formula: opp ~ time + ((0 + t0 | id) + (0 + t1 | id) + (0 + t2 | id) + (0 + t3 | id))## Data: w## ## REML criterion at convergence: 1387.2## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -1.70477 -0.50440 0.02222 0.58653 1.37347 ## ## Random effects:## Groups Name Variance Std.Dev.## id t0 671.8 25.92 ## id.1 t1 478.9 21.88 ## id.2 t2 643.8 25.37 ## id.3 t3 777.7 27.89 ## Residual 625.0 25.00 ## Number of obs: 140, groups: id, 35## ## Fixed effects:## Estimate Std. Error t value## (Intercept) 164.429 5.007 32.839## time 26.925 2.758 9.762## ## Correlation of Fixed Effects:## (Intr)## time -0.801Fully unstructured
We could estimate separate variance and all the covariances - this is just a really complicated model
By default, lme4 will actually try to prevent you from doing this
fully_unstructured <- lmer(opp ~ time + (0 + t0 + t1 + t2 + t3 | id), data = w)## Error: number of observations (=140) <= number of random effects (=140) for term (0 + t0 + t1 + t2 + t3 | id); the random-effects parameters and the residual variance (or scale parameter) are probably unidentifiablearm::display(fully_unstructured)## lmer(formula = opp ~ time + (0 + t0 + t1 + t2 + t3 | id), data = w, ## control = lmerControl(check.nobs.vs.nRE = "ignore"))## coef.est coef.se## (Intercept) 165.83 5.87 ## time 26.58 2.12 ## ## Error terms:## Groups Name Std.Dev. Corr ## id t0 34.42 ## t1 31.58 0.90 ## t2 34.16 0.78 0.94 ## t3 35.95 0.46 0.75 0.88 ## Residual 11.09 ## ---## number of obs: 140, groups: id, 35## AIC = 1289.4, DIC = 1280.3## deviance = 1271.9Terminology
What lme4 fits by default is generally referred to as an unstructured variance-covariance matrix.
This means the random effect variances and covariances are all estimated
The previous model I am referring to as fully unstructured, because we estimate seperate variances at each time point
Some might not make this distinction, so just be careful when people use this term
Autoregressive
Σr=⎡⎢ ⎢ ⎢ ⎢⎣σ2σ2ρσ2ρ2σ2ρ3σ2ρσ2σ2ρσ2ρ2σ2ρ2σ2ρσ2σ2ρσ2ρ3σ2ρ2σ2ρσ2⎤⎥ ⎥ ⎥ ⎥⎦
Each band is forced to be lower than the prior by a constant fraction
- estimated autocorrelation parameter ρ. The error variance is multiplied by ρ for the first diagonal, by ρ2 for the second, etc.
Uses only two variance components
Summary
Notice, you're not estimating random effect variances, just an alternative residual covariance structure
summary(ar)## Generalized least squares fit by REML## Model: opp ~ time ## Data: willett ## AIC BIC logLik## 1281.465 1293.174 -636.7327## ## Correlation Structure: AR(1)## Formula: ~1 | id ## Parameter estimate(s):## Phi ## 0.8249118 ## ## Coefficients:## Value Std.Error t-value p-value## (Intercept) 164.33842 6.136372 26.78104 0## time 27.19786 1.919857 14.16661 0## ## Correlation: ## (Intr)## time -0.469## ## Standardized residuals:## Min Q1 Med Q3 Max ## -2.42825488 -0.71561388 0.03192973 0.78792605 1.76110779 ## ## Residual standard error: 36.37938 ## Degrees of freedom: 140 total; 138 residualExtract composite residual
cm_ar <- corMatrix(ar$modelStruct$corStruct) # all of themcr_ar <- cm_ar[[1]] # just the first (they're all the same)cr_ar## [,1] [,2] [,3] [,4]## [1,] 1.0000000 0.8249118 0.6804795 0.5613356## [2,] 0.8249118 1.0000000 0.8249118 0.6804795## [3,] 0.6804795 0.8249118 1.0000000 0.8249118## [4,] 0.5613356 0.6804795 0.8249118 1.0000000Extract composite residual
cm_ar <- corMatrix(ar$modelStruct$corStruct) # all of themcr_ar <- cm_ar[[1]] # just the first (they're all the same)cr_ar## [,1] [,2] [,3] [,4]## [1,] 1.0000000 0.8249118 0.6804795 0.5613356## [2,] 0.8249118 1.0000000 0.8249118 0.6804795## [3,] 0.6804795 0.8249118 1.0000000 0.8249118## [4,] 0.5613356 0.6804795 0.8249118 1.0000000Multiply the correlation matrix by the model residual variance to get the covariance matrix
cr_ar * sigma(ar)^2## [,1] [,2] [,3] [,4]## [1,] 1323.4596 1091.7375 900.5872 742.9050## [2,] 1091.7375 1323.4596 1091.7375 900.5872## [3,] 900.5872 1091.7375 1323.4596 1091.7375## [4,] 742.9050 900.5872 1091.7375 1323.4596Fit
Note - varIdent specifies different variances for each wave (variances of the identity matrix)
har <- gls( opp ~ time, data = willett, correlation = corAR1(form = ~ 1|id), weights = varIdent(form = ~1|time))Summary
summary(har)## Generalized least squares fit by REML## Model: opp ~ time ## Data: willett ## AIC BIC logLik## 1285.76 1306.25 -635.8798## ## Correlation Structure: AR(1)## Formula: ~1 | id ## Parameter estimate(s):## Phi ## 0.8173622 ## Variance function:## Structure: Different standard deviations per stratum## Formula: ~1 | time ## Parameter estimates:## 0 1 2 3 ## 1.000000 0.915959 0.985068 1.045260 ## ## Coefficients:## Value Std.Error t-value p-value## (Intercept) 164.63344 5.959533 27.62523 0## time 27.11552 1.984807 13.66154 0## ## Correlation: ## (Intr)## time -0.45 ## ## Standardized residuals:## Min Q1 Med Q3 Max ## -2.45009645 -0.74377345 0.02395442 0.82305791 1.81833605 ## ## Residual standard error: 36.17549 ## Degrees of freedom: 140 total; 138 residualExtract/compute composite residual
The below is fairly complicated, but you can work it out if you go line by line
cm_har <- corMatrix(har$modelStruct$corStruct)[[1]] var_struct <- har$modelStruct$varStructvars <- coef(var_struct, unconstrained = FALSE, allCoef = TRUE)vars <- matrix(vars, ncol = 1)cm_har * sigma(har)^2 * (vars %*% t(vars)) # multiply by a mat of vars## [,1] [,2] [,3] [,4]## [1,] 1308.6660 979.7593 861.2399 746.9588## [2,] 979.7593 1097.9457 965.1295 837.0629## [3,] 861.2399 965.1295 1269.8757 1101.3712## [4,] 746.9588 837.0629 1101.3712 1429.8058Summary
summary(toep)## Generalized least squares fit by REML## Model: opp ~ time ## Data: willett ## AIC BIC logLik## 1277.979 1295.543 -632.9896## ## Correlation Structure: ARMA(3,0)## Formula: ~1 | id ## Parameter estimate(s):## Phi1 Phi2 Phi3 ## 0.8039121 0.3665122 -0.3950326 ## ## Coefficients:## Value Std.Error t-value p-value## (Intercept) 165.11855 6.122841 26.96764 0## time 26.91997 2.070391 13.00236 0## ## Correlation: ## (Intr)## time -0.507## ## Standardized residuals:## Min Q1 Med Q3 Max ## -2.44029024 -0.71984566 0.01373249 0.77304950 1.75580973 ## ## Residual standard error: 36.51965 ## Degrees of freedom: 140 total; 138 residualExtract/compute composite residual
Same as with autoregressive - just multiply the correlation matrix by the residual variance
cr_toep <- corMatrix(toep$modelStruct$corStruct)[[1]] cr_toep * sigma(toep)^2## [,1] [,2] [,3] [,4]## [1,] 1333.6848 1105.7350 940.9241 634.8366## [2,] 1105.7350 1333.6848 1105.7350 940.9241## [3,] 940.9241 1105.7350 1333.6848 1105.7350## [4,] 634.8366 940.9241 1105.7350 1333.6848Comparing fits
library(performance)compare_performance(ar, har, toep, w1, varcomp, fully_unstructured, metrics = c("AIC", "BIC"), rank = TRUE) %>% as_tibble()## Warning: Could not get model data.## Warning: Could not get model data.## Warning: Could not get model data.## Warning: When comparing models, please note that probably not all models were fit from same data.## # A tibble: 6 x 5## Name Model AIC BIC Performance_Score## <chr> <chr> <dbl> <dbl> <dbl>## 1 toep gls 1277.979 1295.543 0.9907943## 2 ar gls 1281.465 1293.174 0.9858555## 3 w1 lmerMod 1278.823 1296.473 0.9837572## 4 har gls 1285.760 1306.250 0.9176060## 5 fully_unstructured lmerMod 1289.423 1327.664 0.8195083## 6 varcomp lmerMod 1401.215 1421.806 0We have slight evidence here that the Toeplitz structure fits better than the standard unstructured version, which was slightly better than the autoregressive model.
lme4 models
| Standard | Var Comp | Fully Unstruc | |
|---|---|---|---|
| (Intercept) | 164.374 | 164.429 | 165.832 |
| (6.119) | (5.007) | (5.867) | |
| time | 26.960 | 26.925 | 26.585 |
| (2.167) | (2.758) | (2.121) | |
| sd__(Intercept) | 34.623 | ||
| cor__(Intercept).time | -0.450 | ||
| sd__time | 11.506 | ||
| sd__Observation | 12.629 | 24.999 | 11.087 |
| sd__t0 | 25.919 | 34.424 | |
| sd__t1 | 21.883 | 31.582 | |
| sd__t2 | 25.373 | 34.155 | |
| sd__t3 | 27.887 | 35.947 | |
| cor__t0.t1 | 0.899 | ||
| cor__t0.t2 | 0.784 | ||
| cor__t0.t3 | 0.455 | ||
| cor__t1.t2 | 0.945 | ||
| cor__t1.t3 | 0.754 | ||
| cor__t2.t3 | 0.881 | ||
| AIC | 1278.8 | 1401.2 | 1289.4 |
| BIC | 1296.5 | 1421.8 | 1327.7 |
| Log.Lik. | -633.411 | -693.607 | -631.711 |
| REMLcrit | 1266.823 | 1387.215 | 1263.423 |
Convergence issues
As your models get increasingly complicated, you're likely to run into convergence issues.
Simplifying your residual variance-covariance structure may help
- Keeps the basic model structure intact
Aside - see here for convergence troubleshooting with lme4. The allFit() function is often helpful but very computationally intensive.
Summary
As is probably fairly evident from the code - there are many more structures you could explore. However, most are not implemented through lme4.
Simplifying the residual variance-covariance can sometimes lead to better fitting models
May also be helpful with convergence and avoid overfitting