Modeling Growth 1
Daniel Anderson
Week 5
Model 1
Please translate the following model into lme4::lmer() code
02:00
g5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2(g3_spring),σ2)αj∼N(μαj,σ2αj), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2(g3_spring),σ2)αj∼N(μαj,σ2αj), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,K
Model 1
Please translate the following model into lme4::lmer() code
02:00
g5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2(g3_spring),σ2)αj∼N(μαj,σ2αj), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2(g3_spring),σ2)αj∼N(μαj,σ2αj), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,K
Three equivalent specifications
m1a <- lmer(g5_spring ~ g4_spring + g3_spring + (1|scid) + (1|distid), data = d)m1b <- lmer(g5_spring ~ g4_spring + g3_spring + (1|distid/scid), data = d)m1c <- lmer(g5_spring ~ g4_spring + g3_spring + (1|distid) + (1|distid:scid), data = d)Model 2
Don't worry if you run into convergence warnings
02:00
g5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2j[i](g3_spring),σ2)(αjβ2j)∼N((γα0+γα1(sch_mean_start)μβ2j),(σ2αjραjβ2jρβ2jαjσ2β2j)), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2j[i](g3_spring),σ2)(αjβ2j)∼N((γα0+γα1(sch_mean_start)μβ2j),(σ2αjραjβ2jρβ2jαjσ2β2j)), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,K
Model 2
Don't worry if you run into convergence warnings
02:00
g5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2j[i](g3_spring),σ2)(αjβ2j)∼N((γα0+γα1(sch_mean_start)μβ2j),(σ2αjραjβ2jρβ2jαjσ2β2j)), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1(g4_spring)+β2j[i](g3_spring),σ2)(αjβ2j)∼N((γα0+γα1(sch_mean_start)μβ2j),(σ2αjραjβ2jρβ2jαjσ2β2j)), for scid j = 1,…,Jαk∼N(μαk,σ2αk), for distid k = 1,…,K
lmer(g5_spring ~ g4_spring + g3_spring + sch_mean_start + (g3_spring|scid) + (1|distid), data = d)Model 3
02:00
g5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)(αjβ1jβ2j)∼N((γα0+γα1(sch_mean_start)μβ1jμβ2j),(σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j)), for scid j = 1,…,J(αkβ1kβ2k)∼N((μαkμβ1kμβ2k),(σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k)), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)⎛⎜⎝αjβ1jβ2j⎞⎟⎠∼N⎛⎜ ⎜ ⎜⎝⎛⎜⎝γα0+γα1(sch_mean_start)μβ1jμβ2j⎞⎟⎠,⎛⎜ ⎜ ⎜⎝σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j⎞⎟ ⎟ ⎟⎠⎞⎟ ⎟ ⎟⎠, for scid j = 1,…,J⎛⎜⎝αkβ1kβ2k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝μαkμβ1kμβ2k⎞⎟⎠,⎛⎜ ⎜⎝σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for distid k = 1,…,K
Model 3
02:00
g5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)(αjβ1jβ2j)∼N((γα0+γα1(sch_mean_start)μβ1jμβ2j),(σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j)), for scid j = 1,…,J(αkβ1kβ2k)∼N((μαkμβ1kμβ2k),(σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k)), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)⎛⎜⎝αjβ1jβ2j⎞⎟⎠∼N⎛⎜ ⎜ ⎜⎝⎛⎜⎝γα0+γα1(sch_mean_start)μβ1jμβ2j⎞⎟⎠,⎛⎜ ⎜ ⎜⎝σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j⎞⎟ ⎟ ⎟⎠⎞⎟ ⎟ ⎟⎠, for scid j = 1,…,J⎛⎜⎝αkβ1kβ2k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝μαkμβ1kμβ2k⎞⎟⎠,⎛⎜ ⎜⎝σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for distid k = 1,…,K
lmer(g5_spring ~ g4_spring + g3_spring + sch_mean_start + (g4_spring + g3_spring|scid) + (g4_spring + g3_spring|distid), data = d)Model 4
02:00
g5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)(αjβ1jβ2j)∼N((γα0+γα1(sch_mean_start)μβ1jγβ20+γβ21(sch_mean_start)),(σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j)), for scid j = 1,…,J(αkβ1kβ2k)∼N((μαkμβ1kμβ2k),(σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k)), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)⎛⎜⎝αjβ1jβ2j⎞⎟⎠∼N⎛⎜ ⎜ ⎜⎝⎛⎜ ⎜⎝γα0+γα1(sch_mean_start)μβ1jγβ20+γβ21(sch_mean_start)⎞⎟ ⎟⎠,⎛⎜ ⎜ ⎜⎝σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j⎞⎟ ⎟ ⎟⎠⎞⎟ ⎟ ⎟⎠, for scid j = 1,…,J⎛⎜⎝αkβ1kβ2k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝μαkμβ1kμβ2k⎞⎟⎠,⎛⎜ ⎜⎝σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for distid k = 1,…,K
Model 4
02:00
g5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)(αjβ1jβ2j)∼N((γα0+γα1(sch_mean_start)μβ1jγβ20+γβ21(sch_mean_start)),(σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j)), for scid j = 1,…,J(αkβ1kβ2k)∼N((μαkμβ1kμβ2k),(σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k)), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)⎛⎜⎝αjβ1jβ2j⎞⎟⎠∼N⎛⎜ ⎜ ⎜⎝⎛⎜ ⎜⎝γα0+γα1(sch_mean_start)μβ1jγβ20+γβ21(sch_mean_start)⎞⎟ ⎟⎠,⎛⎜ ⎜ ⎜⎝σ2αjραjβ1jραjβ2jρβ1jαjσ2β1jρβ1jβ2jρβ2jαjρβ2jβ1jσ2β2j⎞⎟ ⎟ ⎟⎠⎞⎟ ⎟ ⎟⎠, for scid j = 1,…,J⎛⎜⎝αkβ1kβ2k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝μαkμβ1kμβ2k⎞⎟⎠,⎛⎜ ⎜⎝σ2αkραkβ1kραkβ2kρβ1kαkσ2β1kρβ1kβ2kρβ2kαkρβ2kβ1kσ2β2k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for distid k = 1,…,K
lmer(g5_spring ~ g4_spring + g3_spring + sch_mean_start + sch_mean_start:g3_spring + (g4_spring + g3_spring|scid) + (g4_spring + g3_spring|distid), data = d)Model 5
A little bit tricky
02:00
g5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)(αjβ1jβ2j)∼N((μαjμβ1jμβ2j),(σ2αj000σ2β1j000σ2β2j)), for scid j = 1,…,J(αkβ1kβ2k)∼N((γα0+γα1(dist_mean_start)μβ1kμβ2k),(σ2αk000σ2β1k000σ2β2k)), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)⎛⎜⎝αjβ1jβ2j⎞⎟⎠∼N⎛⎜ ⎜ ⎜⎝⎛⎜⎝μαjμβ1jμβ2j⎞⎟⎠,⎛⎜ ⎜ ⎜⎝σ2αj000σ2β1j000σ2β2j⎞⎟ ⎟ ⎟⎠⎞⎟ ⎟ ⎟⎠, for scid j = 1,…,J⎛⎜⎝αkβ1kβ2k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝γα0+γα1(dist_mean_start)μβ1kμβ2k⎞⎟⎠,⎛⎜ ⎜⎝σ2αk000σ2β1k000σ2β2k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for distid k = 1,…,K
Model 5
A little bit tricky
02:00
g5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)(αjβ1jβ2j)∼N((μαjμβ1jμβ2j),(σ2αj000σ2β1j000σ2β2j)), for scid j = 1,…,J(αkβ1kβ2k)∼N((γα0+γα1(dist_mean_start)μβ1kμβ2k),(σ2αk000σ2β1k000σ2β2k)), for distid k = 1,…,Kg5_springi∼N(αj[i],k[i]+β1j[i],k[i](g4_spring)+β2j[i],k[i](g3_spring),σ2)⎛⎜⎝αjβ1jβ2j⎞⎟⎠∼N⎛⎜ ⎜ ⎜⎝⎛⎜⎝μαjμβ1jμβ2j⎞⎟⎠,⎛⎜ ⎜ ⎜⎝σ2αj000σ2β1j000σ2β2j⎞⎟ ⎟ ⎟⎠⎞⎟ ⎟ ⎟⎠, for scid j = 1,…,J⎛⎜⎝αkβ1kβ2k⎞⎟⎠∼N⎛⎜ ⎜⎝⎛⎜⎝γα0+γα1(dist_mean_start)μβ1kμβ2k⎞⎟⎠,⎛⎜ ⎜⎝σ2αk000σ2β1k000σ2β2k⎞⎟ ⎟⎠⎞⎟ ⎟⎠, for distid k = 1,…,K
lmer(g5_spring ~ g4_spring + g3_spring + dist_mean_start + (g4_spring + g3_spring||scid) + (g4_spring + g3_spring||distid), data = d)Move to long
l <- d %>% pivot_longer( cols = starts_with("g"), names_to = "timepoint", values_to = "score" )l## # A tibble: 202,500 x 7## # Groups: distid [100]## distid scid sid sch_mean_start dist_mean_start timepoint score## <dbl> <chr> <chr> <dbl> <dbl> <chr> <dbl>## 1 1 1-1 1-1-1 195.1831 189.6743 g3_fall 203.0107## 2 1 1-1 1-1-1 195.1831 189.6743 g3_winter 202.4761## 3 1 1-1 1-1-1 195.1831 189.6743 g3_spring 212.2639## 4 1 1-1 1-1-1 195.1831 189.6743 g4_fall 205.3442## 5 1 1-1 1-1-1 195.1831 189.6743 g4_winter 214.2586## 6 1 1-1 1-1-1 195.1831 189.6743 g4_spring 220.2867## 7 1 1-1 1-1-1 195.1831 189.6743 g5_fall 220.5970## 8 1 1-1 1-1-1 195.1831 189.6743 g5_winter 220.9811## 9 1 1-1 1-1-1 195.1831 189.6743 g5_spring 237.0075## 10 1 1-1 1-1-2 195.1831 189.6743 g3_fall 195.4607## # … with 202,490 more rowsRecode timepoint
First create a data frame that maps the existing values to the new values you want.
wave_frame <- tibble( timepoint = paste0( "g", rep(3:5, each = 3), rep(c("_fall", "_winter", "_spring"), 3) ), wave = 0:8)wave_frame## # A tibble: 9 x 2## timepoint wave## <chr> <int>## 1 g3_fall 0## 2 g3_winter 1## 3 g3_spring 2## 4 g4_fall 3## 5 g4_winter 4## 6 g4_spring 5## 7 g5_fall 6## 8 g5_winter 7## 9 g5_spring 8Join
l <- left_join(l, wave_frame)## Joining, by = "timepoint"l## # A tibble: 202,500 x 8## # Groups: distid [100]## distid scid sid sch_mean_start dist_mean_start timepoint score wave## <dbl> <chr> <chr> <dbl> <dbl> <chr> <dbl> <int>## 1 1 1-1 1-1-1 195.1831 189.6743 g3_fall 203.0107 0## 2 1 1-1 1-1-1 195.1831 189.6743 g3_winter 202.4761 1## 3 1 1-1 1-1-1 195.1831 189.6743 g3_spring 212.2639 2## 4 1 1-1 1-1-1 195.1831 189.6743 g4_fall 205.3442 3## 5 1 1-1 1-1-1 195.1831 189.6743 g4_winter 214.2586 4## 6 1 1-1 1-1-1 195.1831 189.6743 g4_spring 220.2867 5## 7 1 1-1 1-1-1 195.1831 189.6743 g5_fall 220.5970 6## 8 1 1-1 1-1-1 195.1831 189.6743 g5_winter 220.9811 7## 9 1 1-1 1-1-1 195.1831 189.6743 g5_spring 237.0075 8## 10 1 1-1 1-1-2 195.1831 189.6743 g3_fall 195.4607 0## # … with 202,490 more rowsModel 6
02:00
scorei∼N(αj[i],k[i],l[i]+β1j[i],k[i](wave),σ2)(αjβ1j)∼N((μαjμβ1j),(σ2αjραjβ1jρβ1jαjσ2β1j)), for sid j = 1,…,J(αkβ1k)∼N((μαkμβ1k),(σ2αkραkβ1kρβ1kαkσ2β1k)), for scid k = 1,…,Kαl∼N(μαl,σ2αl), for distid l = 1,…,Lscorei∼N(αj[i],k[i],l[i]+β1j[i],k[i](wave),σ2)(αjβ1j)∼N((μαjμβ1j),(σ2αjραjβ1jρβ1jαjσ2β1j)), for sid j = 1,…,J(αkβ1k)∼N((μαkμβ1k),(σ2αkραkβ1kρβ1kαkσ2β1k)), for scid k = 1,…,Kαl∼N(μαl,σ2αl), for distid l = 1,…,L
Model 6
02:00
scorei∼N(αj[i],k[i],l[i]+β1j[i],k[i](wave),σ2)(αjβ1j)∼N((μαjμβ1j),(σ2αjραjβ1jρβ1jαjσ2β1j)), for sid j = 1,…,J(αkβ1k)∼N((μαkμβ1k),(σ2αkραkβ1kρβ1kαkσ2β1k)), for scid k = 1,…,Kαl∼N(μαl,σ2αl), for distid l = 1,…,Lscorei∼N(αj[i],k[i],l[i]+β1j[i],k[i](wave),σ2)(αjβ1j)∼N((μαjμβ1j),(σ2αjραjβ1jρβ1jαjσ2β1j)), for sid j = 1,…,J(αkβ1k)∼N((μαkμβ1k),(σ2αkραkβ1kρβ1kαkσ2β1k)), for scid k = 1,…,Kαl∼N(μαl,σ2αl), for distid l = 1,…,L
lmer(score ~ wave + (wave|sid) + (wave|scid) + (1|distid), data = d)Model 7
This one takes a while to fit, so don't worry about actually fitting it, just try to write the code.
02:00
scorei∼N(αj[i],k[i],l[i]+β1j[i],k[i](wave),σ2)(αjβ1j)∼N((μαjμβ1j),(σ2αjραjβ1jρβ1jαjσ2β1j)), for sid j = 1,…,J(αkβ1k)∼N((γα0+γα1l[i](sch_mean_start)γβ10+γβ11(sch_mean_start)),(σ2αkραkβ1kρβ1kαkσ2β1k)), for scid k = 1,…,K(αlγ1l)∼N((γα0+γα1(dist_mean_start)+γα2(dist_mean_start×wave)μγ1l),(σ2αlραlγ1lργ1lαlσ2γ1l)), for distid l = 1,…,L
Model 7
This one takes a while to fit, so don't worry about actually fitting it, just try to write the code.
02:00
scorei∼N(αj[i],k[i],l[i]+β1j[i],k[i](wave),σ2)(αjβ1j)∼N((μαjμβ1j),(σ2αjραjβ1jρβ1jαjσ2β1j)), for sid j = 1,…,J(αkβ1k)∼N((γα0+γα1l[i](sch_mean_start)γβ10+γβ11(sch_mean_start)),(σ2αkραkβ1kρβ1kαkσ2β1k)), for scid k = 1,…,K(αlγ1l)∼N((γα0+γα1(dist_mean_start)+γα2(dist_mean_start×wave)μγ1l),(σ2αlραlγ1lργ1lαlσ2γ1l)), for distid l = 1,…,L
lmer(score ~ wave + sch_mean_start + dist_mean_start + wave:sch_mean_start + wave:dist_mean_start + (wave|sid) + (wave|scid) + (sch_mean_start|distid), data = l)Look at the data
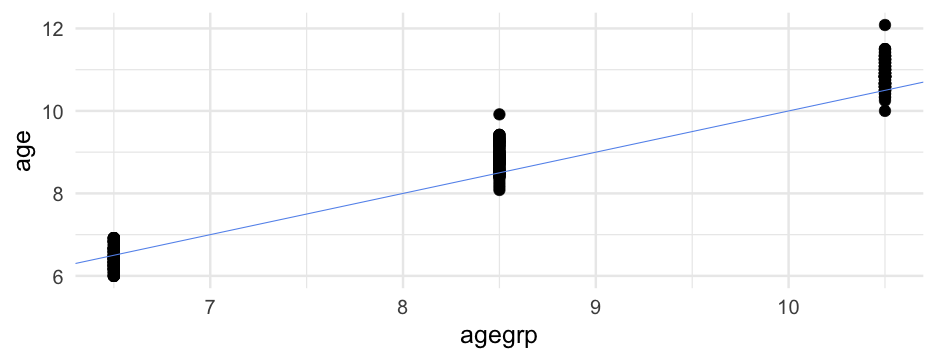
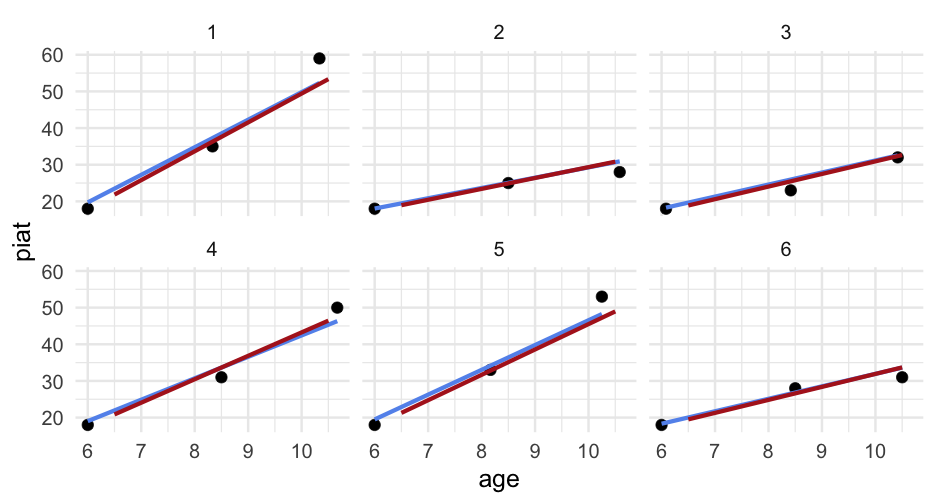
d## # A tibble: 267 x 5## id wave agegrp age piat## <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 1 6.5 6 18## 2 1 2 8.5 8.333333 35## 3 1 3 10.5 10.33333 59## 4 2 1 6.5 6 18## 5 2 2 8.5 8.5 25## 6 2 3 10.5 10.58333 28## 7 3 1 6.5 6.083333 18## 8 3 2 8.5 8.416667 23## 9 3 3 10.5 10.41667 32## 10 4 1 6.5 6 18## # … with 257 more rowsInterpret
arm::display(m_wave)## lmer(formula = piat ~ wave_c + (wave_c | id), data = d)## coef.est coef.se## (Intercept) 21.16 0.62 ## wave_c 10.06 0.59 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 3.38 ## wave_c 4.24 0.22 ## Residual 5.20 ## ---## number of obs: 267, groups: id, 89## AIC = 1830.4, DIC = 1821.5## deviance = 1819.9Interpret
arm::display(m_wave)## lmer(formula = piat ~ wave_c + (wave_c | id), data = d)## coef.est coef.se## (Intercept) 21.16 0.62 ## wave_c 10.06 0.59 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 3.38 ## wave_c 4.24 0.22 ## Residual 5.20 ## ---## number of obs: 267, groups: id, 89## AIC = 1830.4, DIC = 1821.5## deviance = 1819.9But what does a one unit increase in wave_c actually mean?
Interpret
What does the intercept mean here? Age group?
arm::display(m_agegrp)## lmer(formula = piat ~ agegrp + (agegrp | id), data = d, control = lmerControl(optimizer = "bobyqa"))## coef.est coef.se## (Intercept) -11.54 2.21 ## agegrp 5.03 0.30 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 13.43 ## agegrp 2.12 -0.97 ## Residual 5.20 ## ---## number of obs: 267, groups: id, 89## AIC = 1831.8, DIC = 1820.1## deviance = 1819.9Interpret
What does the intercept represent now?
arm::display(m_agegrp2)## lmer(formula = piat ~ agegrp_c + (agegrp_c | id), data = d, control = lmerControl(optimizer = "bobyqa"))## coef.est coef.se## (Intercept) 21.16 0.62 ## agegrp_c 5.03 0.30 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 3.38 ## agegrp_c 2.12 0.22 ## Residual 5.20 ## ---## number of obs: 267, groups: id, 89## AIC = 1831.8, DIC = 1820.1## deviance = 1819.9Interpret
What does the intercept represent now?
arm::display(m_agegrp2)## lmer(formula = piat ~ agegrp_c + (agegrp_c | id), data = d, control = lmerControl(optimizer = "bobyqa"))## coef.est coef.se## (Intercept) 21.16 0.62 ## agegrp_c 5.03 0.30 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 3.38 ## agegrp_c 2.12 0.22 ## Residual 5.20 ## ---## number of obs: 267, groups: id, 89## AIC = 1831.8, DIC = 1820.1## deviance = 1819.9Pop Quiz: Without looking, how do you think the fit of the model has changed?
Comparing fit
library(performance)compare_performance(m_agegrp, m_agegrp2) %>% print_md()Table: Comparison of Model Performance Indices
| Name | Model | AIC | BIC | R2 (cond.) | R2 (marg.) | ICC | RMSE | Sigma |
|---|---|---|---|---|---|---|---|---|
| m_agegrp | lmerMod | 1831.78 | 1853.30 | 0.81 | 0.48 | 0.64 | 4.15 | 5.20 |
| m_agegrp2 | lmerMod | 1831.78 | 1853.30 | 0.81 | 0.48 | 0.64 | 4.15 | 5.20 |
They're identical!
Model assumptions
When we use the
agegrpvariable, we are assuming that all children are the exact same age at each assessment wave.Although
agegrpis more interpretable thanwave, it doesn't solve all our problems You try
Fit another model with age as the time variable instead. How do the results compare?
02:00
Intercept
How do we want to handle this? Probably need to do something. Look at first time point
d %>% filter(wave == 1) %>% count(age)## # A tibble: 12 x 2## age n## <dbl> <int>## 1 6 6## 2 6.083333 4## 3 6.166667 9## 4 6.25 9## 5 6.333333 10## 6 6.416667 11## 7 6.5 7## 8 6.583333 7## 9 6.666667 9## 10 6.75 3## 11 6.833333 7## 12 6.916667 7Summary
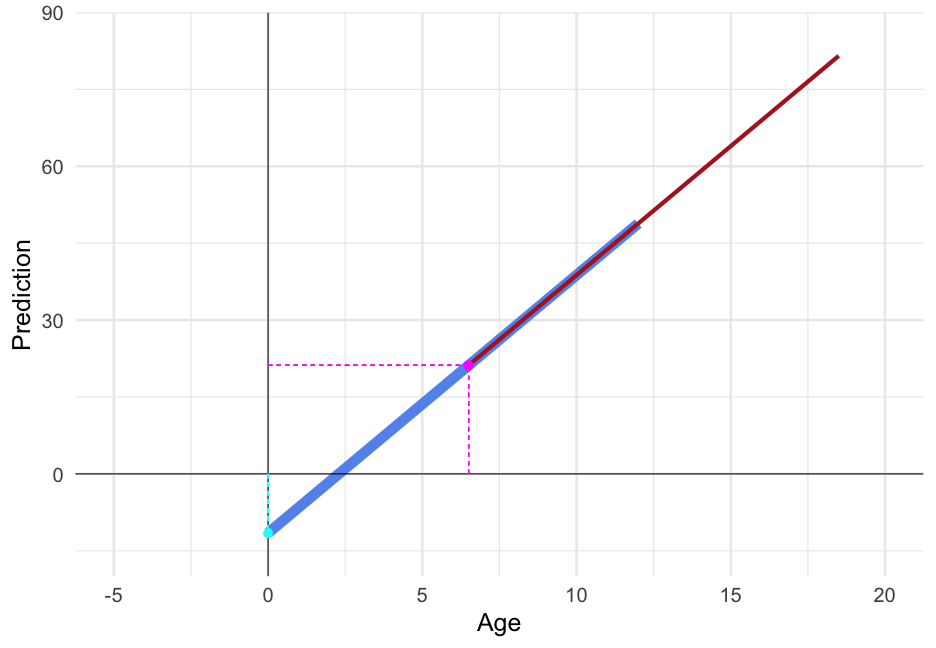
arm::display(m_age)## lmer(formula = piat ~ age6 + (age6 | id), data = d)## coef.est coef.se## (Intercept) 18.79 0.61 ## age6 4.54 0.26 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 2.01 ## age6 1.84 0.17 ## Residual 5.23 ## ---## number of obs: 267, groups: id, 89## AIC = 1816.1, DIC = 1803.6## deviance = 1803.9Compare fit
compare_performance(m_wave, m_agegrp2, m_age) %>% print_md()Table: Comparison of Model Performance Indices
| Name | Model | AIC | BIC | R2 (cond.) | R2 (marg.) | ICC | RMSE | Sigma |
|---|---|---|---|---|---|---|---|---|
| m_wave | lmerMod | 1830.39 | 1851.92 | 0.81 | 0.48 | 0.64 | 4.15 | 5.20 |
| m_agegrp2 | lmerMod | 1831.78 | 1853.30 | 0.81 | 0.48 | 0.64 | 4.15 | 5.20 |
| m_age | lmerMod | 1816.14 | 1837.67 | 0.81 | 0.50 | 0.62 | 4.34 | 5.23 |
Solution
Just multiply age by 12 to get it coded in months.
d <- d %>% mutate(age_months = age6 * 12)d## # A tibble: 267 x 9## id wave agegrp age piat wave_c agegrp_c age6 age_months## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 1 6.5 6 18 0 0 0 0 ## 2 1 2 8.5 8.333333 35 1 2 2.333333 28 ## 3 1 3 10.5 10.33333 59 2 4 4.333333 52 ## 4 2 1 6.5 6 18 0 0 0 0 ## 5 2 2 8.5 8.5 25 1 2 2.5 30 ## 6 2 3 10.5 10.58333 28 2 4 4.583333 55 ## 7 3 1 6.5 6.083333 18 0 0 0.08333333 1.000000## 8 3 2 8.5 8.416667 23 1 2 2.416667 29 ## 9 3 3 10.5 10.41667 32 2 4 4.416667 53 ## 10 4 1 6.5 6 18 0 0 0 0 ## # … with 257 more rowsRefit
m_months <- lmer(piat ~ age_months + (age_months|id), data = d, control = lmerControl(optimizer = "bobyqa"))arm::display(m_months)## lmer(formula = piat ~ age_months + (age_months | id), data = d, ## control = lmerControl(optimizer = "bobyqa"))## coef.est coef.se## (Intercept) 18.79 0.61 ## age_months 0.38 0.02 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 2.01 ## age_months 0.15 0.17 ## Residual 5.23 ## ---## number of obs: 267, groups: id, 89## AIC = 1821.1, DIC = 1798.7## deviance = 1803.9Which model fits better?
Before we test - what do you suspect?
They are not actually the same
compare_performance(m_age, m_months)## # Comparison of Model Performance Indices## ## Name | Model | AIC | BIC | R2 (cond.) | R2 (marg.) | ICC | RMSE | Sigma## ------------------------------------------------------------------------------------------## m_age | lmerMod | 1816.145 | 1837.668 | 0.807 | 0.496 | 0.618 | 4.341 | 5.235## m_months | lmerMod | 1821.115 | 1842.638 | 0.807 | 0.496 | 0.618 | 4.341 | 5.235But they are essentially
pred_frame %>% mutate(pred_months = predict(m_months)[1:18]) %>% select(id, starts_with("pred"))## # A tibble: 18 x 4## id pred_agegrp pred_age pred_months## <dbl> <dbl> <dbl> <dbl>## 1 1 21.85047 19.72314 19.72322## 2 1 37.59817 37.27018 37.27026## 3 1 53.34587 52.31049 52.31057## 4 2 18.95405 18.04288 18.04279## 5 2 24.89376 25.06252 25.06245## 6 2 30.83348 30.91222 30.91217## 7 3 18.88021 18.29429 18.29419## 8 3 25.75976 25.95783 25.95776## 9 3 32.63931 32.52659 32.52655## 10 4 20.86038 19.03189 19.03190## 11 4 33.65203 33.64630 33.64632## 12 4 46.44368 46.31212 46.31215## 13 5 21.28110 19.49879 19.49885## 14 5 35.12409 34.15691 34.15697## 15 5 48.96708 48.25126 48.25132## 16 6 19.51079 18.32752 18.32747## 17 6 26.60084 26.82974 26.82970## 18 6 33.69089 33.63151 33.63148Wages data
Please read in the wages.csv dataset.
wages <- read_csv(here::here("data", "wages.csv"))## ## ── Column specification ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────## cols(## id = col_double(),## lnw = col_double(),## exper = col_double(),## ged = col_double(),## black = col_double(),## hispanic = col_double(),## hgc = col_double(),## uerate = col_double()## )02:00
Data
- Mournane, Boudett, and Willett (1999)
- National Longitudinal Survey of Youth
- Studied wages of individuals who dropped out of high school
Variables
id: Participant IDlnw: Natural log of wagesexper: Experience, in yearsged: Whether or not they completed a GEDblack,hispanic: Dummy variables for race/ethnicityhgc: Highest grade completeduerate: Unemployment rate at the time
Complications
wages %>% filter(id %in% c(206, 332))## # A tibble: 13 x 8## id lnw exper ged black hispanic hgc uerate## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 206 2.028 1.874 0 0 0 10 9.2 ## 2 206 2.297 2.814 0 0 0 10 11 ## 3 206 2.482 4.314 0 0 0 10 6.295## 4 332 1.63 0.125 0 0 1 8 7.1 ## 5 332 1.476 1.625 0 0 1 8 9.6 ## 6 332 1.804 2.413 0 0 1 8 7.2 ## 7 332 1.439 3.393 0 0 1 8 6.195## 8 332 1.748 4.47 0 0 1 8 5.595## 9 332 1.526 5.178 0 0 1 8 4.595## 10 332 2.044 6.082 0 0 1 8 4.295## 11 332 2.179 7.043 0 0 1 8 3.395## 12 332 2.186 8.197 0 0 1 8 4.395## 13 332 4.035 9.092 0 0 1 8 6.695arm::display(m_wage0)## lmer(formula = lnw ~ exper + (exper | id), data = wages, control = lmerControl(optimizer = "bobyqa"))## coef.est coef.se## (Intercept) 1.72 0.01 ## exper 0.05 0.00 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 0.23 ## exper 0.04 -0.30 ## Residual 0.31 ## ---## number of obs: 6402, groups: id, 888## AIC = 4951.3, DIC = 4903.5## deviance = 4921.4arm::display(m_wage0)## lmer(formula = lnw ~ exper + (exper | id), data = wages, control = lmerControl(optimizer = "bobyqa"))## coef.est coef.se## (Intercept) 1.72 0.01 ## exper 0.05 0.00 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 0.23 ## exper 0.04 -0.30 ## Residual 0.31 ## ---## number of obs: 6402, groups: id, 888## AIC = 4951.3, DIC = 4903.5## deviance = 4921.4Every one year of extra experience corresponded to a 0.05 increase in log wages, on average, which varied across participants with a standard deviation of 0.04.
Race/Ethnicity
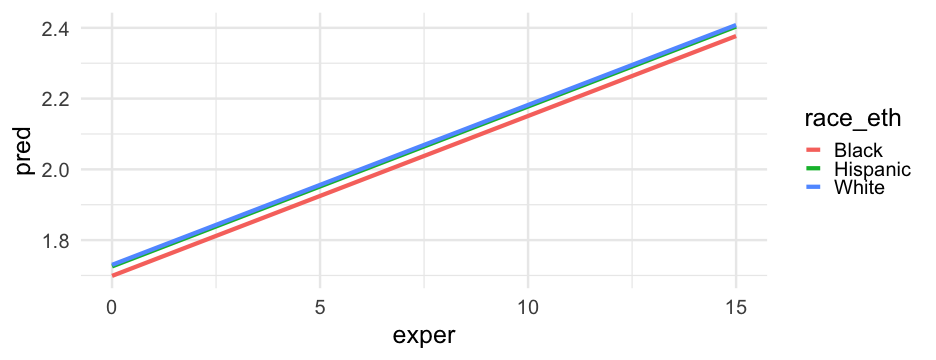
Let's create a new variable that has all the race/ethnicity labels instead of the dummy codes.
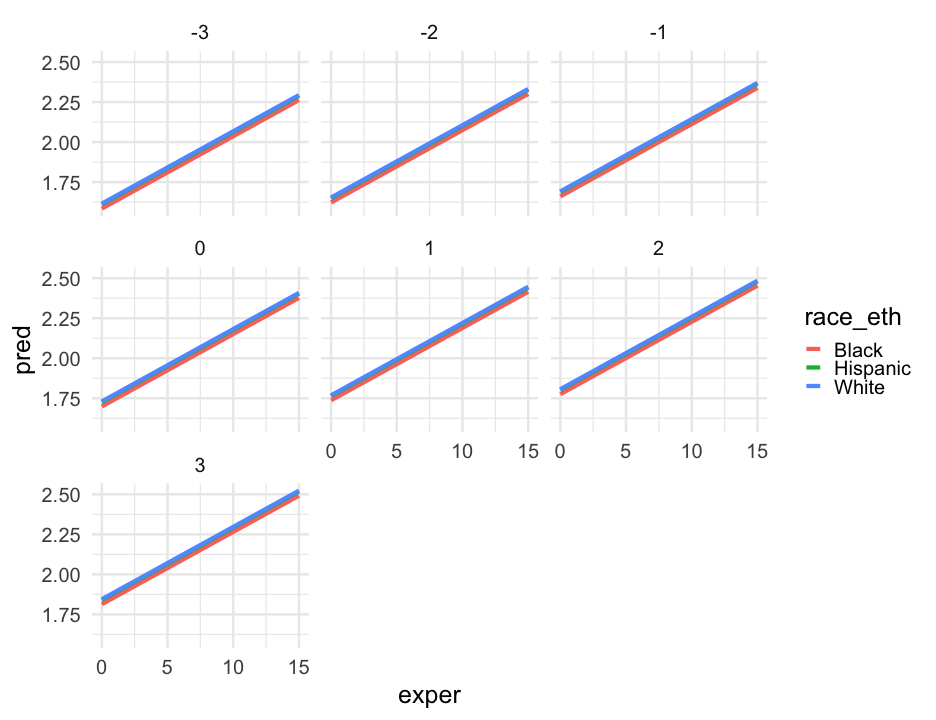
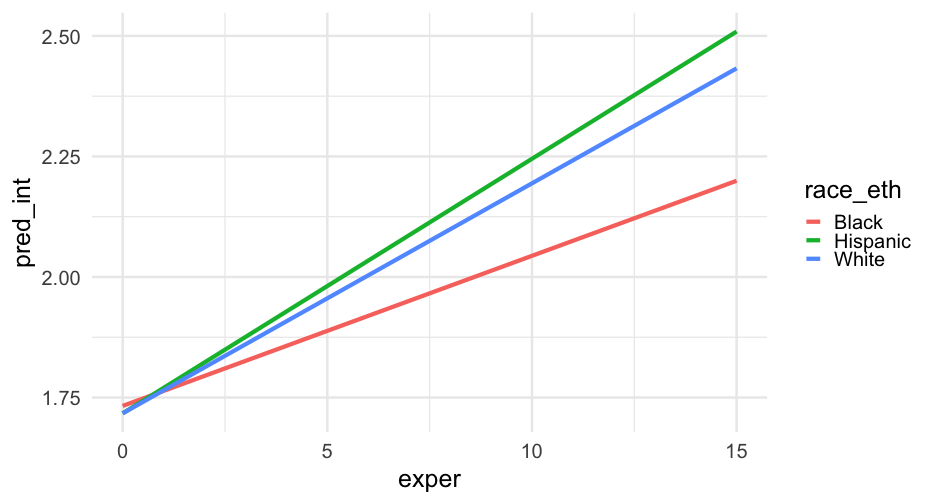
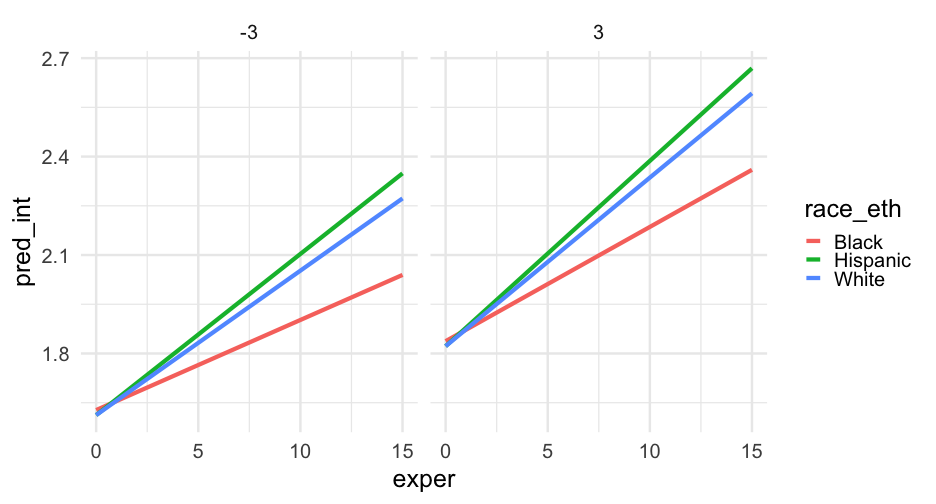
pred_frame <- pred_frame %>% mutate( race_eth = case_when( black == 0 & hispanic == 0 ~ "White", black == 1 & hispanic == 0 ~ "Black", black == 0 & hispanic == 1 ~ "Hispanic", TRUE ~ NA_character_ ) )Interactions
If we want to know how the slope may or may not depend on these variables, we have to model the interactions.
Just the two-way interactions
m_wage2 <- lmer(lnw ~ exper + black + exper:black + exper:hispanic + hgc_9 + exper:hgc_9 + (exper|id), data = wages, control = lmerControl(optimizer = "bobyqa"))Focus on hgc
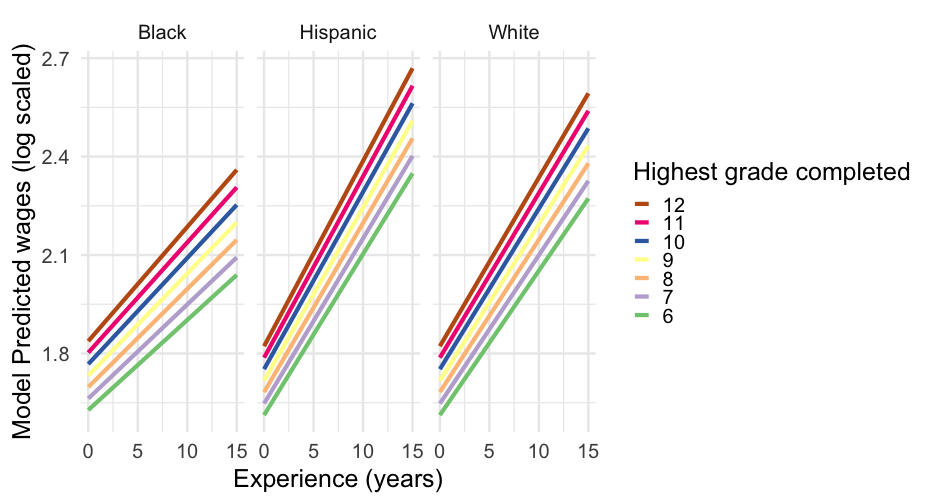
pred_frame %>% drop_na() %>% ggplot(aes(exper, pred_int)) + geom_line(aes(color = factor(hgc_9))) + facet_wrap(~race_eth) + scale_color_brewer("Highest grade completed", palette = "Accent", breaks = 3:-3, labels = 12:6) + labs(x = "Experience (years)", y = "Model Predicted wages (log scaled)")Model summary
arm::display(m_wage2)## lmer(formula = lnw ~ exper + black + exper:black + exper:hispanic + ## hgc_9 + exper:hgc_9 + (exper | id), data = wages, control = lmerControl(optimizer = "bobyqa"))## coef.est coef.se## (Intercept) 1.72 0.01 ## exper 0.05 0.00 ## black 0.02 0.02 ## hgc_9 0.03 0.01 ## exper:black -0.02 0.01 ## exper:hispanic 0.01 0.00 ## exper:hgc_9 0.00 0.00 ## ## Error terms:## Groups Name Std.Dev. Corr ## id (Intercept) 0.23 ## exper 0.04 -0.31 ## Residual 0.31 ## ---## number of obs: 6402, groups: id, 888## AIC = 4955.1, DIC = 4811.9## deviance = 4872.5The data
Simulated data to mimic a common form of non-linearity.
Notice the "true" intercept and slope for each student is actually in the data.
sim_d <- read_csv(here::here("data", "curvilinear-sim.csv"))sim_d## # A tibble: 2,760 x 5## sid int slope date score## <dbl> <dbl> <dbl> <date> <dbl>## 1 1 31.91237 32.25614 2019-04-26 116.1638 ## 2 1 31.91237 32.25614 2019-04-14 50.02688## 3 1 31.91237 32.25614 2019-05-21 151.8375 ## 4 2 22.91502 24.05294 2019-04-25 81.93698## 5 2 22.91502 24.05294 2019-04-30 93.47374## 6 2 22.91502 24.05294 2019-05-24 113.8067 ## 7 2 22.91502 24.05294 2019-04-27 87.83396## 8 2 22.91502 24.05294 2019-05-29 112.8697 ## 9 2 22.91502 24.05294 2019-04-25 82.40156## 10 2 22.91502 24.05294 2019-05-27 111.8477 ## # … with 2,750 more rowsFit linear model
linear <- lmer(score ~ days_from_start + (days_from_start|sid), data = sim_d, control = lmerControl(optimizer = "Nelder_Mead"))arm::display(linear)## lmer(formula = score ~ days_from_start + (days_from_start | sid), ## data = sim_d, control = lmerControl(optimizer = "Nelder_Mead"))## coef.est coef.se## (Intercept) 68.66 0.71 ## days_from_start 1.22 0.02 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 13.69 ## days_from_start 0.28 -0.41 ## Residual 7.91 ## ---## number of obs: 2760, groups: sid, 500## AIC = 21005, DIC = 20981.9## deviance = 20987.4Quadratic summary
arm::display(quad)## lmer(formula = score ~ days_from_start + days2 + (days_from_start | ## sid), data = sim_d, control = lmerControl(optimizer = "Nelder_Mead"))## coef.est coef.se## (Intercept) 52.09 0.54 ## days_from_start 2.98 0.03 ## days2 -0.03 0.00 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 10.07 ## days_from_start 0.18 0.31 ## Residual 4.41 ## ---## number of obs: 2760, groups: sid, 500## AIC = 18279.8, DIC = 18224.7## deviance = 18245.2Compare
anova(linear, quad)## refitting model(s) with ML (instead of REML)## Data: sim_d## Models:## linear: score ~ days_from_start + (days_from_start | sid)## quad: score ~ days_from_start + days2 + (days_from_start | sid)## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) ## linear 6 20999 21035 -10493.7 20987 ## quad 7 18259 18301 -9122.6 18245 2742.2 1 < 2.2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Plot predictions
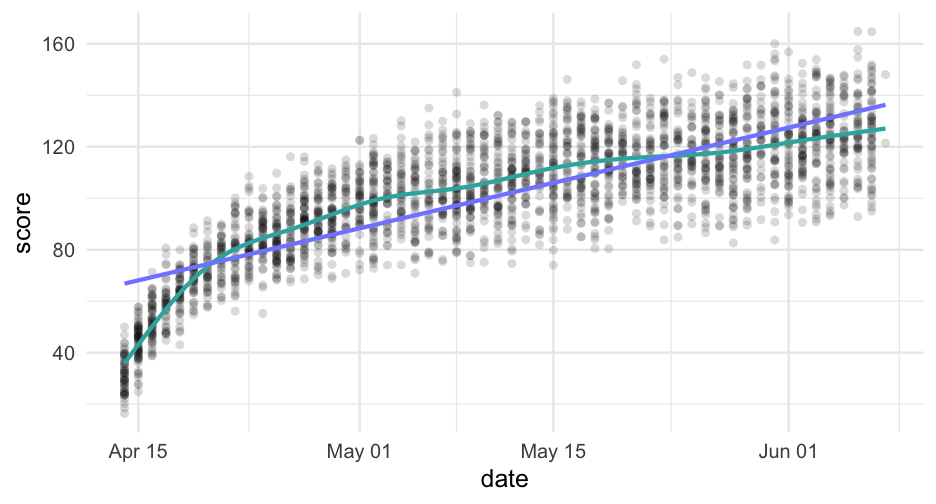
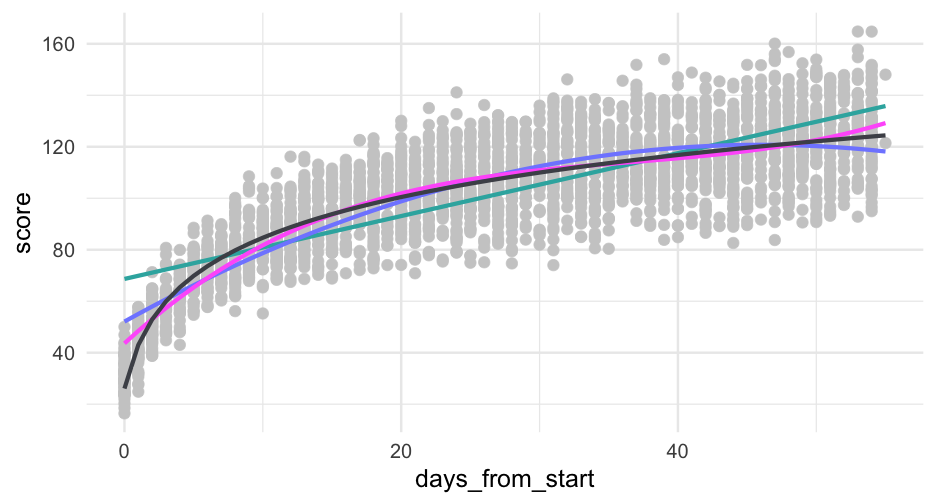
pred_frame <- tibble( days_from_start = 0:max(sim_d$days_from_start), days2 = days_from_start^2, sid = -999 ) %>% mutate(pred_linear = predict(linear, newdata = ., allow.new.levels = TRUE), pred_quad = predict(quad, newdata = ., allow.new.levels = TRUE))pred_frame## # A tibble: 56 x 5## days_from_start days2 sid pred_linear pred_quad## <int> <dbl> <dbl> <dbl> <dbl>## 1 0 0 -999 68.65771 52.09304## 2 1 1 -999 69.87886 55.04428## 3 2 4 -999 71.10000 57.93071## 4 3 9 -999 72.32114 60.75233## 5 4 16 -999 73.54228 63.50914## 6 5 25 -999 74.76342 66.20114## 7 6 36 -999 75.98456 68.82834## 8 7 49 -999 77.20570 71.39072## 9 8 64 -999 78.42684 73.88829## 10 9 81 -999 79.64798 76.32105## # … with 46 more rowsggplot(pred_frame, aes(days_from_start)) + geom_point(aes(y = score), data = sim_d, color = "gray80") + geom_line(aes(y = pred_linear), color = "#33B1AE") + geom_line(aes(y = pred_quad), color = "#808AFF")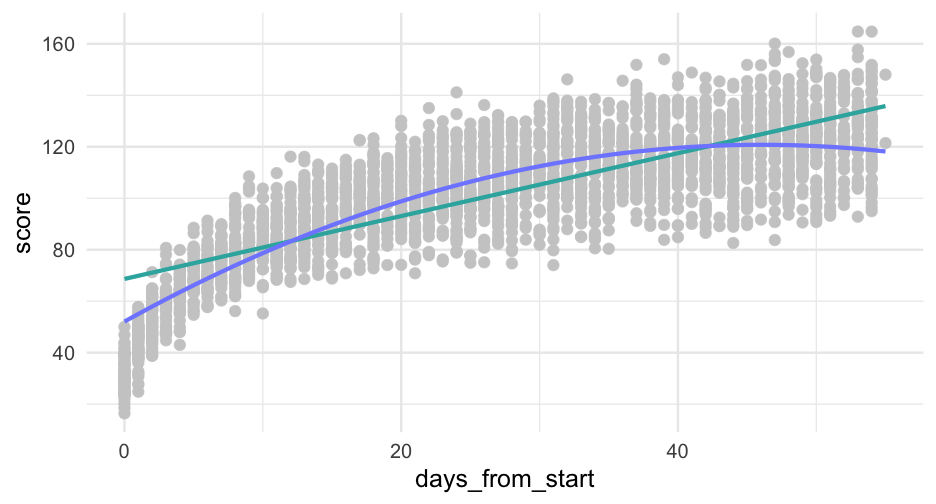
This is definitely looking better, but it's too high in the lower tail and maybe a bit too low in the upper
Cubic?
You try first - extend what we just did to model a cubic trend
03:00
sim_d <- sim_d %>% mutate(days3 = days_from_start^3)cubic <- lmer(score ~ days_from_start + days2 + days3 + (days_from_start|sid), data = sim_d, control = lmerControl(optimizer = "Nelder_Mead"))## Warning: Some predictor variables are on very different scales: consider rescalingCubic summary
arm::display(cubic)## lmer(formula = score ~ days_from_start + days2 + days3 + (days_from_start | ## sid), data = sim_d, control = lmerControl(optimizer = "Nelder_Mead"))## coef.est coef.se## (Intercept) 43.64 0.49 ## days_from_start 4.93 0.04 ## days2 -0.12 0.00 ## days3 0.00 0.00 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 9.48 ## days_from_start 0.15 0.55 ## Residual 2.81 ## ---## number of obs: 2760, groups: sid, 500## AIC = 16311.2, DIC = 16211.2## deviance = 16253.2Compare
anova(linear, quad, cubic)## refitting model(s) with ML (instead of REML)## Data: sim_d## Models:## linear: score ~ days_from_start + (days_from_start | sid)## quad: score ~ days_from_start + days2 + (days_from_start | sid)## cubic: score ~ days_from_start + days2 + days3 + (days_from_start | ## cubic: sid)## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) ## linear 6 20999 21035 -10493.7 20987 ## quad 7 18259 18301 -9122.6 18245 2742.2 1 < 2.2e-16 ***## cubic 8 16269 16317 -8126.6 16253 1992.0 1 < 2.2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Predictions
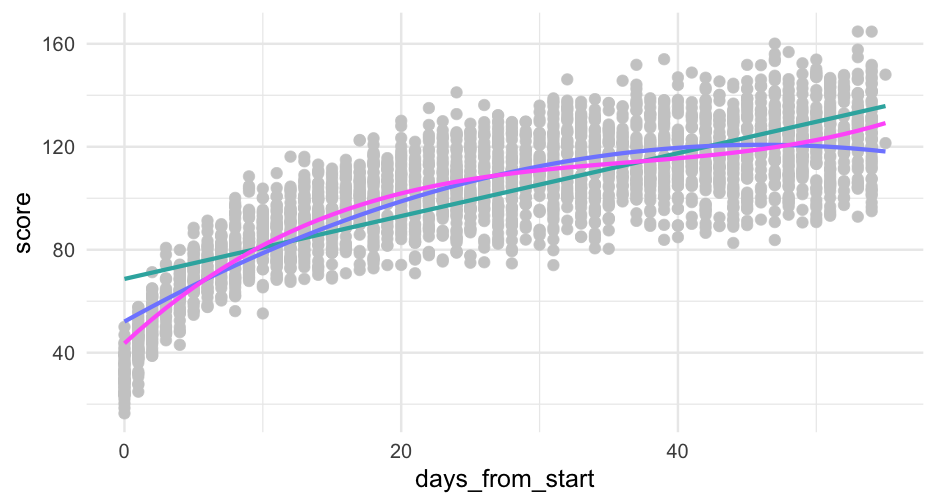
pred_frame <- pred_frame %>% mutate(days3 = days_from_start^3)pred_frame %>% mutate(pred_cubic = predict(cubic, newdata = ., allow.new.levels = TRUE)) %>% ggplot(aes(days_from_start)) + geom_point(aes(y = score), data = sim_d, color = "gray80") + geom_line(aes(y = pred_linear), color = "#33B1AE") + geom_line(aes(y = pred_quad), color = "#808AFF") + geom_line(aes(y = pred_cubic), color = "#ff66fa")Try log
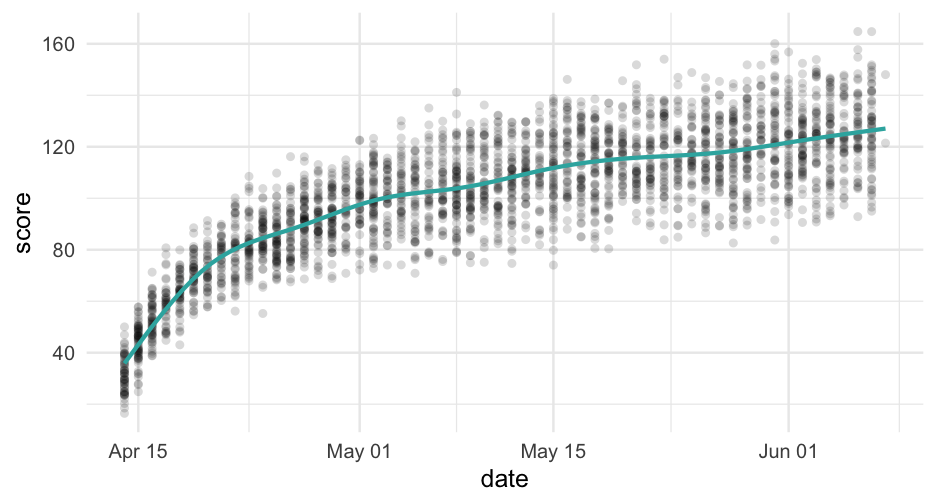
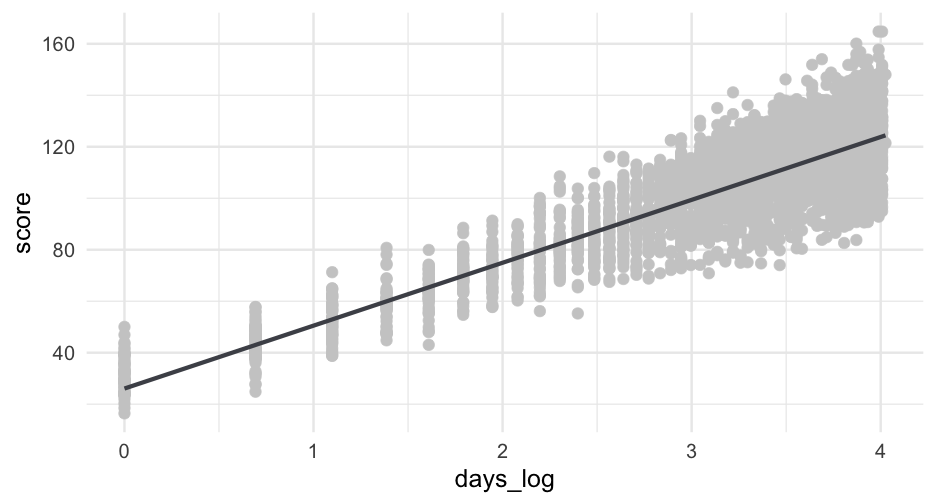
If you're familiar with log growth, the scatterplots we've been looking at probably resemble this trend quite well.
Let's try log transforming our time variable, then fit with it - bonus, we save two estimated parameters.
Note - I will have to use log(x + 1) instead of log(x) because log(0) =−∞ and log(1) =0.
sim_d <- sim_d %>% mutate(days_log = log(days_from_start + 1))log_m <- lmer(score ~ days_log + (days_log|sid), data = sim_d)arm::display(log_m)## lmer(formula = score ~ days_log + (days_log | sid), data = sim_d)## coef.est coef.se## (Intercept) 26.08 0.32 ## days_log 24.43 0.15 ## ## Error terms:## Groups Name Std.Dev. Corr ## sid (Intercept) 6.16 ## days_log 3.05 0.20 ## Residual 1.52 ## ---## number of obs: 2760, groups: sid, 500## AIC = 13703.9, DIC = 13687## deviance = 13689.5Compare
anova(linear, quad, cubic, log_m)## refitting model(s) with ML (instead of REML)## Data: sim_d## Models:## linear: score ~ days_from_start + (days_from_start | sid)## log_m: score ~ days_log + (days_log | sid)## quad: score ~ days_from_start + days2 + (days_from_start | sid)## cubic: score ~ days_from_start + days2 + days3 + (days_from_start | ## cubic: sid)## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) ## linear 6 20999 21035 -10493.7 20987 ## log_m 6 13702 13737 -6844.7 13690 7298 0 ## quad 7 18259 18301 -9122.6 18245 0 1 1 ## cubic 8 16269 16317 -8126.6 16253 1992 1 <2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1It use the same number of parameters as the linear model, but fits far better.
On raw scale
pred_frame %>% mutate(pred_cubic = predict(cubic, newdata = ., allow.new.levels = TRUE)) %>% ggplot(aes(days_from_start)) + geom_point(aes(y = score), data = sim_d, color = "gray80") + geom_line(aes(y = pred_linear), color = "#33B1AE") + geom_line(aes(y = pred_quad), color = "#808AFF") + geom_line(aes(y = pred_cubic), color = "#ff66fa") + geom_line(aes(y = pred_log), color = "#4D4F57")