Full applications w/Bayes
Plus some ways of handling missing data
Daniel Anderson
Week 8
Data
nurses## # A tibble: 1,000 x 11## hospital ward wardid nurse age gender experien stress wardtype hospsize expcon ## <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr> <chr> ## 1 1 1 11 1 36 Male 11 7 general care large experiment## 2 1 1 11 2 45 Male 20 7 general care large experiment## 3 1 1 11 3 32 Male 7 7 general care large experiment## 4 1 1 11 4 57 Female 25 6 general care large experiment## 5 1 1 11 5 46 Female 22 6 general care large experiment## 6 1 1 11 6 60 Female 22 6 general care large experiment## 7 1 1 11 7 23 Female 13 6 general care large experiment## 8 1 1 11 8 32 Female 13 7 general care large experiment## 9 1 1 11 9 60 Male 17 7 general care large experiment## 10 1 2 12 10 45 Male 21 6 special care large experiment## # … with 990 more rowsModel 1
Fit the following model
$$ \small \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i]} + \beta_{1j[i]}(\operatorname{experien}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\gamma^{\beta_{1}}_{0} + \gamma^{\beta_{1}}_{1}(\operatorname{wardtype}_{\operatorname{special\ care}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \end{aligned} $$
02:00
Model 1
Fit the following model
$$ \small \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i]} + \beta_{1j[i]}(\operatorname{experien}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\gamma^{\beta_{1}}_{0} + \gamma^{\beta_{1}}_{1}(\operatorname{wardtype}_{\operatorname{special\ care}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \end{aligned} $$
02:00
lmer(stress ~ experien * wardtype + (experien|wardid), data = nurses)# orlmer(stress ~ experien + wardtype + experien:wardtype + (experien|wardid), data = nurses)Model 2
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{experien}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\gamma^{\beta_{1}}_{0} + \gamma^{\beta_{1}}_{1}(\operatorname{wardtype}_{\operatorname{special\ care}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\mu_{\beta_{1k}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{1k}} \\ \rho_{\beta_{1k}\alpha_{k}} & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
Model 2
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{experien}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\gamma^{\beta_{1}}_{0} + \gamma^{\beta_{1}}_{1}(\operatorname{wardtype}_{\operatorname{special\ care}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\mu_{\beta_{1k}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{1k}} \\ \rho_{\beta_{1k}\alpha_{k}} & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
lmer(stress ~ experien * wardtype + hospsize + (experien|wardid) + (experien|hospital), data = nurses)Model 3
Fit the following model
$$ \small \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{experien}) + \beta_{2}(\operatorname{age}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{expcon}_{\operatorname{experiment}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & 0 \\ 0 & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\mu_{\alpha_{k}} \\ &\mu_{\beta_{1k}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & 0 \\ 0 & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
Model 3
Fit the following model
$$ \small \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{experien}) + \beta_{2}(\operatorname{age}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{expcon}_{\operatorname{experiment}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & 0 \\ 0 & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\mu_{\alpha_{k}} \\ &\mu_{\beta_{1k}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & 0 \\ 0 & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
lmer(stress ~ experien + age + expcon + (experien||wardid) + (experien||hospital), data = nurses)# orlmer(stress ~ experien + age + expcon + (1|wardid) + (0 + experien|wardid) + (1|hospital) + (0 + experien|hospital), data = nurses)Model 4
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{experien}) + \beta_{2}(\operatorname{age}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{expcon}_{\operatorname{experiment}}) + \gamma_{2}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\gamma^{\beta_{1}}_{0} + \gamma^{\beta_{1}}_{1}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma^{\beta_{1}}_{2}(\operatorname{hospsize}_{\operatorname{small}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{1k}} \\ \rho_{\beta_{1k}\alpha_{k}} & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
Model 4
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{stress}_{i} &\sim N \left(\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{experien}) + \beta_{2}(\operatorname{age}), \sigma^2 \right) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{expcon}_{\operatorname{experiment}}) + \gamma_{2}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\gamma^{\beta_{1}}_{0} + \gamma^{\beta_{1}}_{1}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma^{\beta_{1}}_{2}(\operatorname{hospsize}_{\operatorname{small}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{1k}} \\ \rho_{\beta_{1k}\alpha_{k}} & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
lmer(stress ~ experien * hospsize + age + expcon + wardtype + (experien|wardid) + (experien|hospital), data = nurses)Model 5
Fit the following model
$$ \small \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1j[i]}(\operatorname{age}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\mu_{\alpha_{j}} \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \alpha_{k} &\sim N \left(\mu_{\alpha_{k}}, \sigma^2_{\alpha_{k}} \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
03:00
Model 5
Fit the following model
$$ \small \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1j[i]}(\operatorname{age}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\mu_{\alpha_{j}} \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \alpha_{k} &\sim N \left(\mu_{\alpha_{k}}, \sigma^2_{\alpha_{k}} \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
03:00
nurses <- nurses %>% mutate(expcon = factor(expcon))glmer(expcon ~ age + (age|wardid) + (1|hospital), data = nurses, family = binomial(link = "logit"), data = nurses)Model 6
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1j[i]}(\operatorname{age}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \alpha_{k} &\sim N \left(\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}), \sigma^2_{\alpha_{k}} \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
Model 6
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1j[i]}(\operatorname{age}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \alpha_{k} &\sim N \left(\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}), \sigma^2_{\alpha_{k}} \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
glmer(expcon ~ hospsize + age + wardtype + (age|wardid) + (1|hospital), data = nurses, family = binomial(link = "logit"))Model 7
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{age}) + \beta_{2}(\operatorname{gender}_{\operatorname{Male}}) + \beta_{3}(\operatorname{age} \times \operatorname{gender}_{\operatorname{Male}}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\mu_{\beta_{1k}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{1k}} \\ \rho_{\beta_{1k}\alpha_{k}} & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
Model 7
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1j[i],k[i]}(\operatorname{age}) + \beta_{2}(\operatorname{gender}_{\operatorname{Male}}) + \beta_{3}(\operatorname{age} \times \operatorname{gender}_{\operatorname{Male}}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{1j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\mu_{\beta_{1j}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{1j}} \\ \rho_{\beta_{1j}\alpha_{j}} & \sigma^2_{\beta_{1j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{1k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\mu_{\beta_{1k}} \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{1k}} \\ \rho_{\beta_{1k}\alpha_{k}} & \sigma^2_{\beta_{1k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
glmer(expcon ~ age * gender + wardtype + hospsize + (age|wardid) + (age|hospital), data = nurses, family = binomial(link = "logit"))Model 8
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1}(\operatorname{age}) + \beta_{2}(\operatorname{gender}_{\operatorname{Male}}) + \beta_{3j[i],k[i]}(\operatorname{experien}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{3j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\gamma^{\beta_{3}}_{0} + \gamma^{\beta_{3}}_{1}(\operatorname{wardtype}_{\operatorname{special\ care}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{3j}} \\ \rho_{\beta_{3j}\alpha_{j}} & \sigma^2_{\beta_{3j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{3k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\gamma^{\beta_{3}}_{0} + \gamma^{\beta_{3}}_{1}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma^{\beta_{3}}_{2}(\operatorname{hospsize}_{\operatorname{small}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{3k}} \\ \rho_{\beta_{3k}\alpha_{k}} & \sigma^2_{\beta_{3k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
Model 8
Fit the following model
$$ \scriptsize \begin{aligned} \operatorname{expcon}_{i} &\sim \operatorname{Binomial}(n = 1, \operatorname{prob}_{\operatorname{expcon} = \operatorname{experiment}} = \widehat{P}) \\ \log\left[\frac{\hat{P}}{1 - \hat{P}} \right] &=\alpha_{j[i],k[i]} + \beta_{1}(\operatorname{age}) + \beta_{2}(\operatorname{gender}_{\operatorname{Male}}) + \beta_{3j[i],k[i]}(\operatorname{experien}) \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{j} \\ &\beta_{3j} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{wardtype}_{\operatorname{special\ care}}) \\ &\gamma^{\beta_{3}}_{0} + \gamma^{\beta_{3}}_{1}(\operatorname{wardtype}_{\operatorname{special\ care}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{j}} & \rho_{\alpha_{j}\beta_{3j}} \\ \rho_{\beta_{3j}\alpha_{j}} & \sigma^2_{\beta_{3j}} \end{array} \right) \right) \text{, for wardid j = 1,} \dots \text{,J} \\ \left( \begin{array}{c} \begin{aligned} &\alpha_{k} \\ &\beta_{3k} \end{aligned} \end{array} \right) &\sim N \left( \left( \begin{array}{c} \begin{aligned} &\gamma_{0}^{\alpha} + \gamma_{1}^{\alpha}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma_{2}^{\alpha}(\operatorname{hospsize}_{\operatorname{small}}) \\ &\gamma^{\beta_{3}}_{0} + \gamma^{\beta_{3}}_{1}(\operatorname{hospsize}_{\operatorname{medium}}) + \gamma^{\beta_{3}}_{2}(\operatorname{hospsize}_{\operatorname{small}}) \end{aligned} \end{array} \right) , \left( \begin{array}{cc} \sigma^2_{\alpha_{k}} & \rho_{\alpha_{k}\beta_{3k}} \\ \rho_{\beta_{3k}\alpha_{k}} & \sigma^2_{\beta_{3k}} \end{array} \right) \right) \text{, for hospital k = 1,} \dots \text{,K} \end{aligned} $$
02:00
glmer(expcon ~ age + gender + experien * wardtype + experien * hospsize + (experien|wardid) + (experien|hospital), data = nurses, family = binomial(link = "logit"))The data
Twitter!
Real data, collected Wednesday, but anonymized
You can see the code I used to get the data, but you'll pull different data if you run it
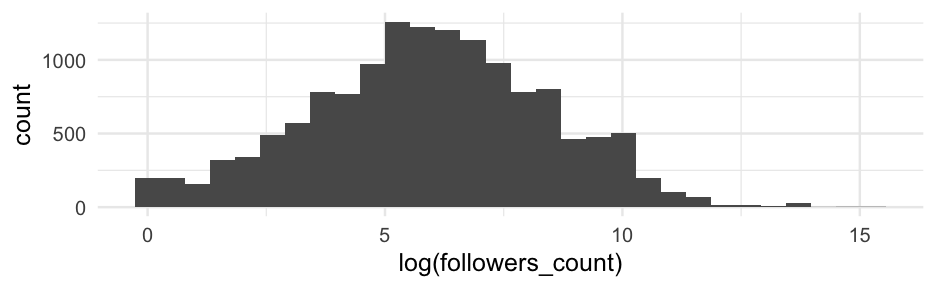
18,000 tweets including the hashtag #blm
Sentence-level text coded for sentiment using the {sentimentr} package, then averaged for the entire tweet
Read in the data
It's a little different because there's still a list column of hashtags. Use code like the following:
library(tidyverse)blm <- read_rds(here::here("data", "blm_sentiment.Rds"))blm## # A tibble: 14,339 x 21## user_id status_id trump_in_description followers_count friends_count listed_count statuses_count favourites_count account_created_at verified## <dbl> <dbl> <lgl> <int> <int> <int> <int> <int> <date> <lgl> ## 1 1691 14339 FALSE 964 859 16 22975 60680 2020-05-24 FALSE ## 2 7740 14338 FALSE 135 389 2 10624 1027 2009-07-25 FALSE ## 3 313 14337 FALSE 261 31 1 154 54 2013-01-06 FALSE ## 4 5740 14336 FALSE 4032 3872 23 23101 57150 2014-08-22 FALSE ## 5 5740 2927 FALSE 4032 3872 23 23101 57150 2014-08-22 FALSE ## 6 5740 9379 FALSE 4032 3872 23 23101 57150 2014-08-22 FALSE ## 7 5740 2457 FALSE 4032 3872 23 23101 57150 2014-08-22 FALSE ## 8 5740 7854 FALSE 4032 3872 23 23101 57150 2014-08-22 FALSE ## 9 5740 1550 FALSE 4032 3872 23 23101 57150 2014-08-22 FALSE ## 10 5740 11789 FALSE 4032 3872 23 23101 57150 2014-08-22 FALSE ## # … with 14,329 more rows, and 11 more variables: tweet_created_at <dttm>, word_count <int>, is_reply <lgl>, is_quote <lgl>, has_url <lgl>, has_photo <lgl>,## # n_mentions <int>, hashtags <list>, favorite_count <int>, retweet_count <int>, is_positive_sentiment <int>Getting more info
This is a data frame like any other with one exception - the hashtags column is a list.
See all hashtags
blm %>% unnest(hashtags) %>% count(hashtags, sort = TRUE) # %>%## # A tibble: 12,011 x 2## hashtags n## <chr> <int>## 1 BLM 12209## 2 blm 2231## 3 BlackLivesMatter 2153## 4 GeorgeFloyd 1473## 5 SashaJohnson 559## 6 racism 416## 7 blacklivesmatter 410## 8 LGBTQ 360## 9 FBR 291## 10 Resist 289## # … with 12,001 more rows# View()List column
We can unnest() to see all of them, but we can't use that in modeling
Pull more features
Let's get the number of hashtags in the tweet
blm <- blm %>% rowwise() %>% mutate(n_hashtags = length(hashtags)) %>% ungroup() blm %>% select(user_id, n_hashtags)## # A tibble: 14,339 x 2## user_id n_hashtags## <dbl> <int>## 1 1691 1## 2 7740 1## 3 313 16## 4 5740 5## 5 5740 5## 6 5740 7## 7 5740 5## 8 5740 7## 9 5740 7## 10 5740 5## # … with 14,329 more rowsWhat about trump_in_description?
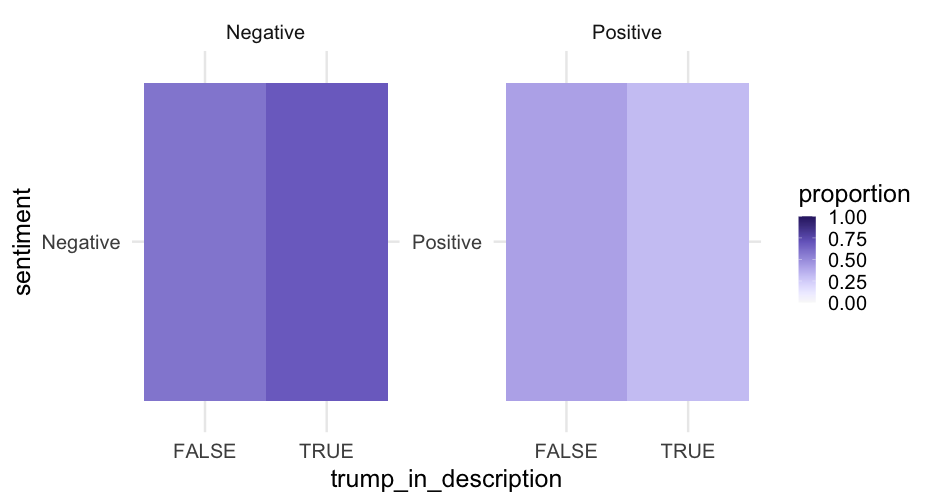
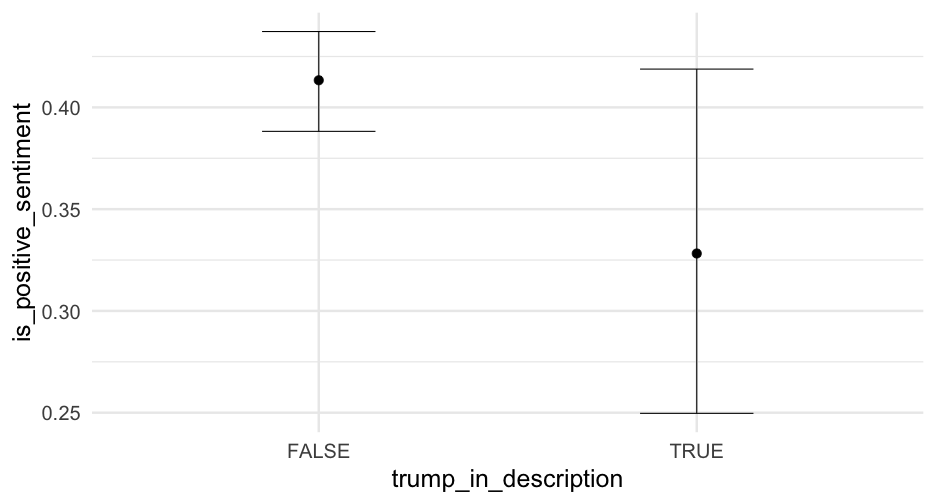
trump_proportions <- blm %>% mutate(sentiment = ifelse( is_positive_sentiment > 0, "Positive", "Negative" ) ) %>% count(trump_in_description, sentiment) %>% group_by(trump_in_description) %>% mutate(proportion = n/sum(n))trump_proportions## # A tibble: 4 x 4## # Groups: trump_in_description [2]## trump_in_description sentiment n proportion## <lgl> <chr> <int> <dbl>## 1 FALSE Negative 8130 0.5848921## 2 FALSE Positive 5770 0.4151079## 3 TRUE Negative 301 0.6856492## 4 TRUE Positive 138 0.3143508Account creation
In reality I would probably explore my data for a bit longer, unless I already knew a lot about out. For now, let's just do one more, looking at the relation between when their account was created, and whether the sentiment was positive.
ggplot(blm, aes(account_created_at, factor(is_positive_sentiment))) + geom_jitter(width = 0, alpha = 0.05)
Last bit
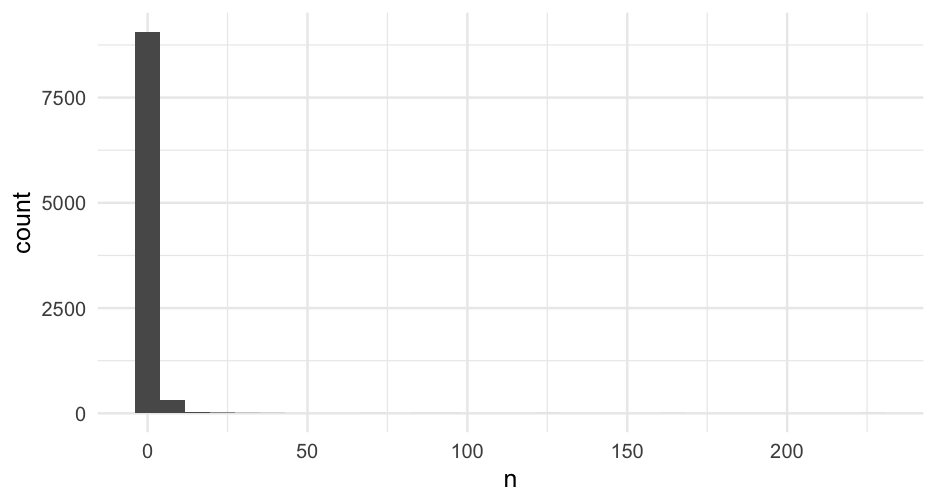
Sample size issues
The number of tweets per person varies a lot.
When \(n = 1\) it's not theoretically a problem, although I've had issues with estimation in the past.
You might consider including the number of tweets a person as a predictor.
- Could be an indicator they are a bot or a journalist.
See here for more information
Maximum likelihood version
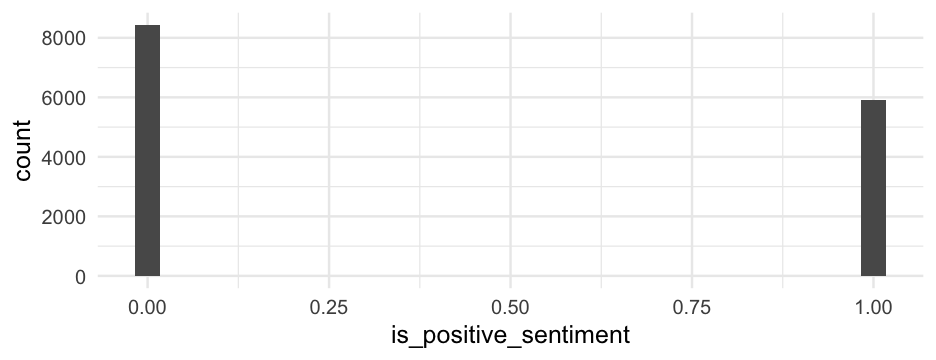
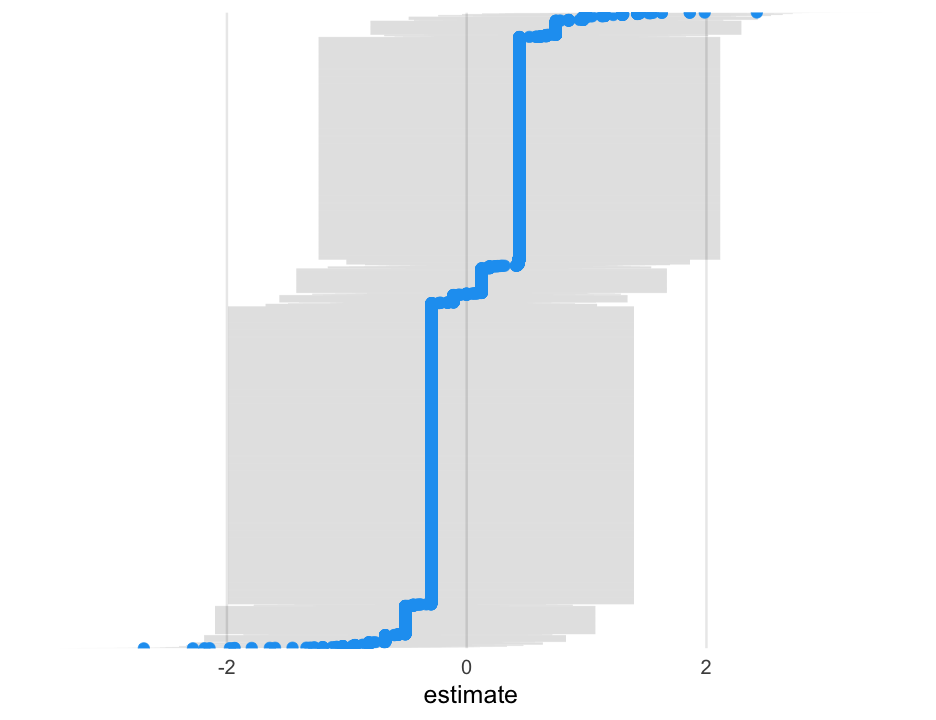
library(lme4)m0_ml <- glmer(is_positive_sentiment ~ 1 + (1|user_id), data = blm, family = binomial(link = "logit"))arm::display(m0_ml)## glmer(formula = is_positive_sentiment ~ 1 + (1 | user_id), data = blm, ## family = binomial(link = "logit"))## coef.est coef.se ## -0.40 0.02 ## ## Error terms:## Groups Name Std.Dev.## user_id (Intercept) 0.95 ## Residual 1.00 ## ---## number of obs: 14339, groups: user_id, 9454## AIC = 18757.7, DIC = 10514.1## deviance = 14633.9Variability
The log-odds varied between people with a standard deviation of 0.95.
The probability of a person one standard deviation above and below the average posting a positive tweet were estimated at:
# Probability for an individual 1 SD belowbrms::inv_logit_scaled(-0.40 - 0.95)## [1] 0.2058704# Probability for an individual 1 SD abovebrms::inv_logit_scaled(-0.40 + 0.95)## [1] 0.6341356Plot the variability
First pull the random effect estimates (deviations from the fixed effect)
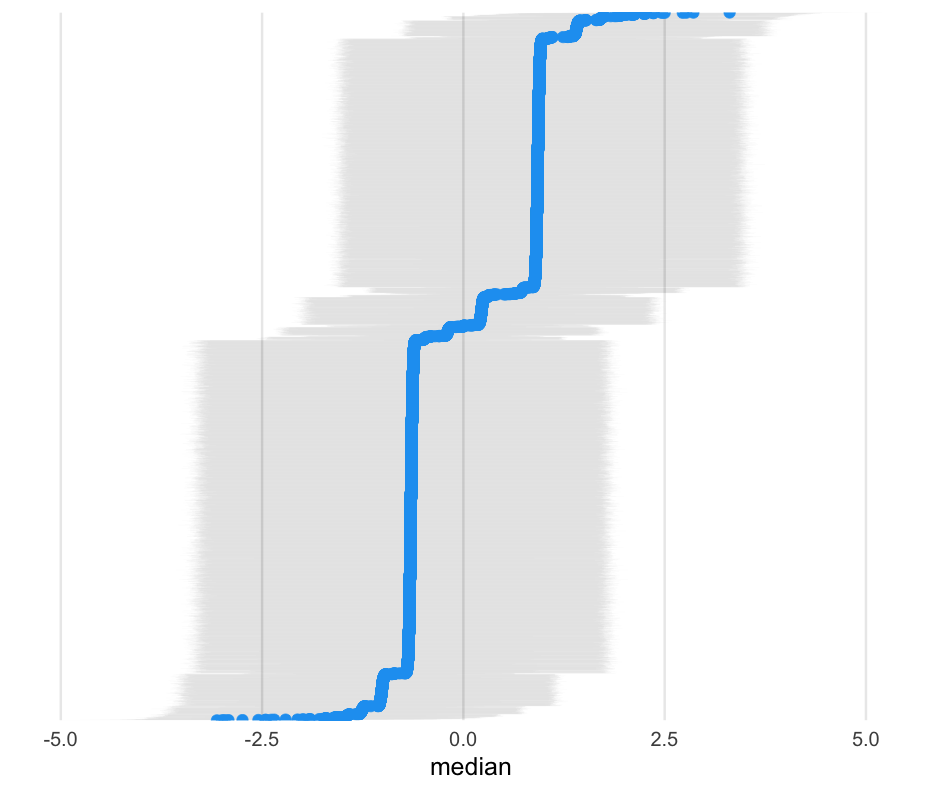
library(broom.mixed)tidy_m0_ml <- tidy(m0_ml, "ran_vals", conf.int = TRUE) %>% mutate(level = fct_reorder(level, estimate))Next create the plot
ggplot(tidy_m0_ml, aes(estimate, level)) + geom_linerange(aes(xmin = conf.low, xmax = conf.high), alpha = 0.01) + geom_point(color = "#1DA1F2") + # get rid of some plot elements theme(axis.text.y = element_blank(), axis.title.y = element_blank(), panel.grid.major.y = element_blank(), panel.grid.minor = element_blank())Fit using Bayes
You try first. Fit the same model using Bayes. Go ahead and assume flat priors.
04:00
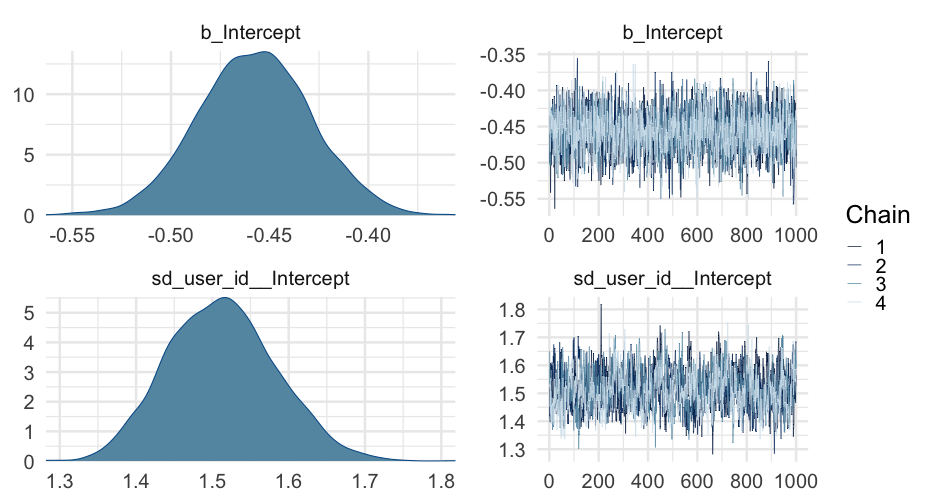
library(brms)m0_b <- brm(is_positive_sentiment ~ 1 + (1|user_id), data = blm, family = bernoulli(link = "logit"), cores = 4, backend = "cmdstanr")## -\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/Running MCMC with 4 parallel chains...## ## Chain 1 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 2 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 3 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 4 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 2 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 4 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 1 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 3 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 2 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 4 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 1 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 3 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 2 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 1 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 4 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 3 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 2 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 1 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 4 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 3 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 2 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 1 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 4 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 3 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 2 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 1 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 4 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 3 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 2 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 1 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 4 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 3 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 2 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 1 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 4 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 3 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 2 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 1 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 4 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 3 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 2 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 2 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 1 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 1 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 4 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 4 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 3 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 3 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 2 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 1 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 4 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 3 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 2 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 1 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 4 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 3 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 2 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 1 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 4 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 3 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 2 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 1 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 4 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 3 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 2 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 1 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 4 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 3 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 2 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 1 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 4 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 3 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 2 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 1 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 4 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 3 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 2 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 1 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 4 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 3 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 2 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 1 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 4 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 3 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 2 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 2 finished in 112.4 seconds.## Chain 1 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 1 finished in 113.4 seconds.## Chain 4 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 4 finished in 114.1 seconds.## Chain 3 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 3 finished in 115.0 seconds.## ## All 4 chains finished successfully.## Mean chain execution time: 113.7 seconds.## Total execution time: 115.4 seconds.Summary
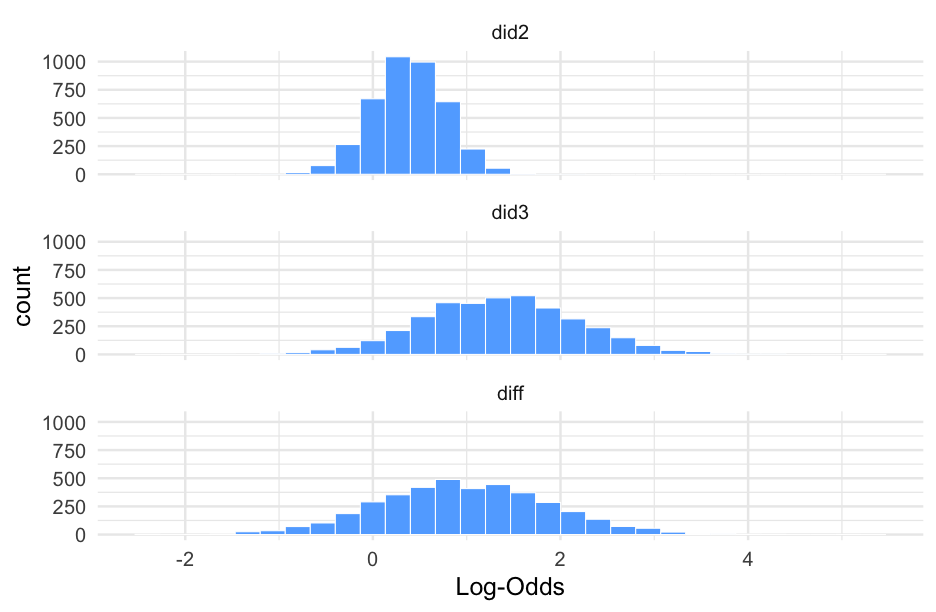
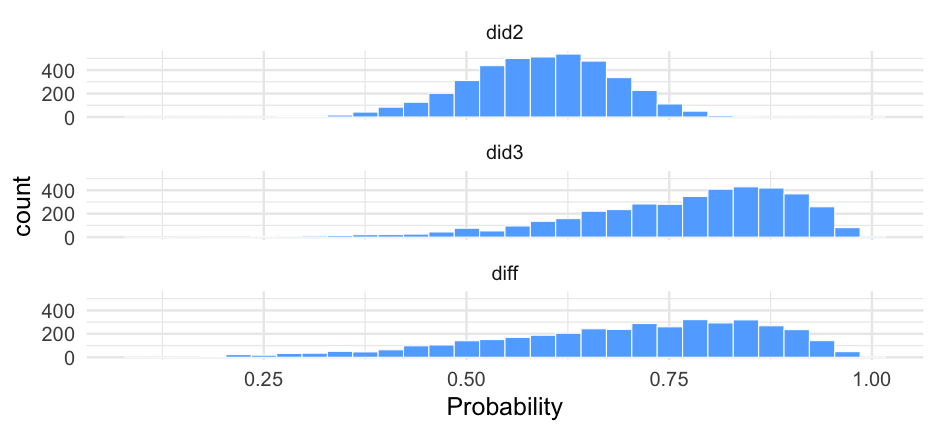
Notice the variance is fairly different here
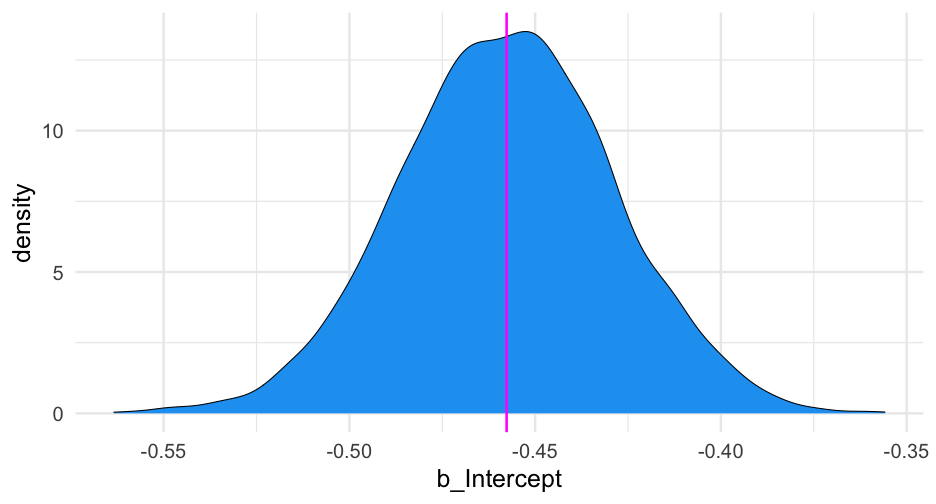
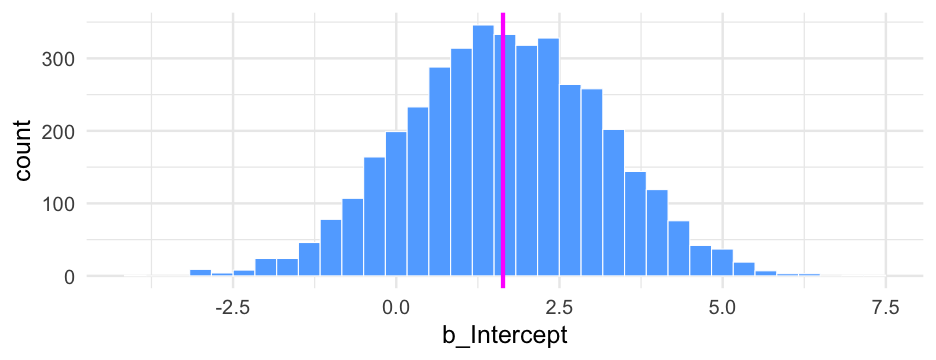
summary(m0_b)## Family: bernoulli ## Links: mu = logit ## Formula: is_positive_sentiment ~ 1 + (1 | user_id) ## Data: blm (Number of observations: 14339) ## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;## total post-warmup samples = 4000## ## Group-Level Effects: ## ~user_id (Number of levels: 9454) ## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS## sd(Intercept) 1.51 0.07 1.38 1.65 1.01 528 1224## ## Population-Level Effects: ## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS## Intercept -0.46 0.03 -0.51 -0.40 1.00 2643 2906## ## Samples were drawn using sample(hmc). For each parameter, Bulk_ESS## and Tail_ESS are effective sample size measures, and Rhat is the potential## scale reduction factor on split chains (at convergence, Rhat = 1).Plot person-estimates
We have to go to {tidybayes} for this
General purpose tool to pull lots of different things from our model and plot them
For now, we'll do the plotting ourselves
Let's start by looking at what's actually in the model
In this case r_* implies "random". These are the deviations from the average.
library(tidybayes)get_variables(m0_b)## [1] "b_Intercept" "sd_user_id__Intercept" "Intercept" "r_user_id[1,Intercept]" "r_user_id[2,Intercept]" ## [6] "r_user_id[3,Intercept]" "r_user_id[4,Intercept]" "r_user_id[5,Intercept]" "r_user_id[6,Intercept]" "r_user_id[7,Intercept]" ## [11] "r_user_id[8,Intercept]" "r_user_id[9,Intercept]" "r_user_id[10,Intercept]" "r_user_id[11,Intercept]" "r_user_id[12,Intercept]" ## [16] "r_user_id[13,Intercept]" "r_user_id[14,Intercept]" "r_user_id[15,Intercept]" "r_user_id[16,Intercept]" "r_user_id[17,Intercept]" ## [21] "r_user_id[18,Intercept]" "r_user_id[19,Intercept]" "r_user_id[20,Intercept]" "r_user_id[21,Intercept]" "r_user_id[22,Intercept]" ## [26] "r_user_id[23,Intercept]" "r_user_id[24,Intercept]" "r_user_id[25,Intercept]" "r_user_id[26,Intercept]" "r_user_id[27,Intercept]" ## [31] "r_user_id[28,Intercept]" "r_user_id[29,Intercept]" "r_user_id[30,Intercept]" "r_user_id[31,Intercept]" "r_user_id[32,Intercept]" ## [36] "r_user_id[33,Intercept]" "r_user_id[34,Intercept]" "r_user_id[35,Intercept]" "r_user_id[36,Intercept]" "r_user_id[37,Intercept]" ## [41] "r_user_id[38,Intercept]" "r_user_id[39,Intercept]" "r_user_id[40,Intercept]" "r_user_id[41,Intercept]" "r_user_id[42,Intercept]" ## [46] "r_user_id[43,Intercept]" "r_user_id[44,Intercept]" "r_user_id[45,Intercept]" "r_user_id[46,Intercept]" "r_user_id[47,Intercept]" ## [51] "r_user_id[48,Intercept]" "r_user_id[49,Intercept]" "r_user_id[50,Intercept]" "r_user_id[51,Intercept]" "r_user_id[52,Intercept]" ## [56] "r_user_id[53,Intercept]" "r_user_id[54,Intercept]" "r_user_id[55,Intercept]" "r_user_id[56,Intercept]" "r_user_id[57,Intercept]" ## [61] "r_user_id[58,Intercept]" "r_user_id[59,Intercept]" "r_user_id[60,Intercept]" "r_user_id[61,Intercept]" "r_user_id[62,Intercept]" ## [66] "r_user_id[63,Intercept]" "r_user_id[64,Intercept]" "r_user_id[65,Intercept]" "r_user_id[66,Intercept]" "r_user_id[67,Intercept]" ## [71] "r_user_id[68,Intercept]" "r_user_id[69,Intercept]" "r_user_id[70,Intercept]" "r_user_id[71,Intercept]" "r_user_id[72,Intercept]" ## [76] "r_user_id[73,Intercept]" "r_user_id[74,Intercept]" "r_user_id[75,Intercept]" "r_user_id[76,Intercept]" "r_user_id[77,Intercept]" ## [81] "r_user_id[78,Intercept]" "r_user_id[79,Intercept]" "r_user_id[80,Intercept]" "r_user_id[81,Intercept]" "r_user_id[82,Intercept]" ## [86] "r_user_id[83,Intercept]" "r_user_id[84,Intercept]" "r_user_id[85,Intercept]" "r_user_id[86,Intercept]" "r_user_id[87,Intercept]" ## [91] "r_user_id[88,Intercept]" "r_user_id[89,Intercept]" "r_user_id[90,Intercept]" "r_user_id[91,Intercept]" "r_user_id[92,Intercept]" ## [96] "r_user_id[93,Intercept]" "r_user_id[94,Intercept]" "r_user_id[95,Intercept]" "r_user_id[96,Intercept]" "r_user_id[97,Intercept]" ## [101] "r_user_id[98,Intercept]" "r_user_id[99,Intercept]" "r_user_id[100,Intercept]" "r_user_id[101,Intercept]" "r_user_id[102,Intercept]"## [106] "r_user_id[103,Intercept]" "r_user_id[104,Intercept]" "r_user_id[105,Intercept]" "r_user_id[106,Intercept]" "r_user_id[107,Intercept]"## [111] "r_user_id[108,Intercept]" "r_user_id[109,Intercept]" "r_user_id[110,Intercept]" "r_user_id[111,Intercept]" "r_user_id[112,Intercept]"## [116] "r_user_id[113,Intercept]" "r_user_id[114,Intercept]" "r_user_id[115,Intercept]" "r_user_id[116,Intercept]" "r_user_id[117,Intercept]"## [121] "r_user_id[118,Intercept]" "r_user_id[119,Intercept]" "r_user_id[120,Intercept]" "r_user_id[121,Intercept]" "r_user_id[122,Intercept]"## [126] "r_user_id[123,Intercept]" "r_user_id[124,Intercept]" "r_user_id[125,Intercept]" "r_user_id[126,Intercept]" "r_user_id[127,Intercept]"## [131] "r_user_id[128,Intercept]" "r_user_id[129,Intercept]" "r_user_id[130,Intercept]" "r_user_id[131,Intercept]" "r_user_id[132,Intercept]"## [136] "r_user_id[133,Intercept]" "r_user_id[134,Intercept]" "r_user_id[135,Intercept]" "r_user_id[136,Intercept]" "r_user_id[137,Intercept]"## [141] "r_user_id[138,Intercept]" "r_user_id[139,Intercept]" "r_user_id[140,Intercept]" "r_user_id[141,Intercept]" "r_user_id[142,Intercept]"## [146] "r_user_id[143,Intercept]" "r_user_id[144,Intercept]" "r_user_id[145,Intercept]" "r_user_id[146,Intercept]" "r_user_id[147,Intercept]"## [151] "r_user_id[148,Intercept]" "r_user_id[149,Intercept]" "r_user_id[150,Intercept]" "r_user_id[151,Intercept]" "r_user_id[152,Intercept]"## [156] "r_user_id[153,Intercept]" "r_user_id[154,Intercept]" "r_user_id[155,Intercept]" "r_user_id[156,Intercept]" "r_user_id[157,Intercept]"## [161] "r_user_id[158,Intercept]" "r_user_id[159,Intercept]" "r_user_id[160,Intercept]" "r_user_id[161,Intercept]" "r_user_id[162,Intercept]"## [166] "r_user_id[163,Intercept]" "r_user_id[164,Intercept]" "r_user_id[165,Intercept]" "r_user_id[166,Intercept]" "r_user_id[167,Intercept]"## [171] "r_user_id[168,Intercept]" "r_user_id[169,Intercept]" "r_user_id[170,Intercept]" "r_user_id[171,Intercept]" "r_user_id[172,Intercept]"## [176] "r_user_id[173,Intercept]" "r_user_id[174,Intercept]" "r_user_id[175,Intercept]" "r_user_id[176,Intercept]" "r_user_id[177,Intercept]"## [181] "r_user_id[178,Intercept]" "r_user_id[179,Intercept]" "r_user_id[180,Intercept]" "r_user_id[181,Intercept]" "r_user_id[182,Intercept]"## [186] "r_user_id[183,Intercept]" "r_user_id[184,Intercept]" "r_user_id[185,Intercept]" "r_user_id[186,Intercept]" "r_user_id[187,Intercept]"## [191] "r_user_id[188,Intercept]" "r_user_id[189,Intercept]" "r_user_id[190,Intercept]" "r_user_id[191,Intercept]" "r_user_id[192,Intercept]"## [196] "r_user_id[193,Intercept]" "r_user_id[194,Intercept]" "r_user_id[195,Intercept]" "r_user_id[196,Intercept]" "r_user_id[197,Intercept]"## [201] "r_user_id[198,Intercept]" "r_user_id[199,Intercept]" "r_user_id[200,Intercept]" "r_user_id[201,Intercept]" "r_user_id[202,Intercept]"## [206] "r_user_id[203,Intercept]" "r_user_id[204,Intercept]" "r_user_id[205,Intercept]" "r_user_id[206,Intercept]" "r_user_id[207,Intercept]"## [211] "r_user_id[208,Intercept]" "r_user_id[209,Intercept]" "r_user_id[210,Intercept]" "r_user_id[211,Intercept]" "r_user_id[212,Intercept]"## [216] "r_user_id[213,Intercept]" "r_user_id[214,Intercept]" "r_user_id[215,Intercept]" "r_user_id[216,Intercept]" "r_user_id[217,Intercept]"## [221] "r_user_id[218,Intercept]" "r_user_id[219,Intercept]" "r_user_id[220,Intercept]" "r_user_id[221,Intercept]" "r_user_id[222,Intercept]"## [226] "r_user_id[223,Intercept]" "r_user_id[224,Intercept]" "r_user_id[225,Intercept]" "r_user_id[226,Intercept]" "r_user_id[227,Intercept]"## [231] "r_user_id[228,Intercept]" "r_user_id[229,Intercept]" "r_user_id[230,Intercept]" "r_user_id[231,Intercept]" "r_user_id[232,Intercept]"## [236] "r_user_id[233,Intercept]" "r_user_id[234,Intercept]" "r_user_id[235,Intercept]" "r_user_id[236,Intercept]" "r_user_id[237,Intercept]"## [241] "r_user_id[238,Intercept]" "r_user_id[239,Intercept]" "r_user_id[240,Intercept]" "r_user_id[241,Intercept]" "r_user_id[242,Intercept]"## [246] "r_user_id[243,Intercept]" "r_user_id[244,Intercept]" "r_user_id[245,Intercept]" "r_user_id[246,Intercept]" "r_user_id[247,Intercept]"## [251] "r_user_id[248,Intercept]" "r_user_id[249,Intercept]" "r_user_id[250,Intercept]" "r_user_id[251,Intercept]" "r_user_id[252,Intercept]"## [256] "r_user_id[253,Intercept]" "r_user_id[254,Intercept]" "r_user_id[255,Intercept]" "r_user_id[256,Intercept]" "r_user_id[257,Intercept]"## [261] "r_user_id[258,Intercept]" "r_user_id[259,Intercept]" "r_user_id[260,Intercept]" "r_user_id[261,Intercept]" "r_user_id[262,Intercept]"## [266] "r_user_id[263,Intercept]" "r_user_id[264,Intercept]" "r_user_id[265,Intercept]" "r_user_id[266,Intercept]" "r_user_id[267,Intercept]"## [271] "r_user_id[268,Intercept]" "r_user_id[269,Intercept]" "r_user_id[270,Intercept]" "r_user_id[271,Intercept]" "r_user_id[272,Intercept]"## [276] "r_user_id[273,Intercept]" "r_user_id[274,Intercept]" "r_user_id[275,Intercept]" "r_user_id[276,Intercept]" "r_user_id[277,Intercept]"## [281] "r_user_id[278,Intercept]" "r_user_id[279,Intercept]" "r_user_id[280,Intercept]" "r_user_id[281,Intercept]" "r_user_id[282,Intercept]"## [286] "r_user_id[283,Intercept]" "r_user_id[284,Intercept]" "r_user_id[285,Intercept]" "r_user_id[286,Intercept]" "r_user_id[287,Intercept]"## [291] "r_user_id[288,Intercept]" "r_user_id[289,Intercept]" "r_user_id[290,Intercept]" "r_user_id[291,Intercept]" "r_user_id[292,Intercept]"## [296] "r_user_id[293,Intercept]" "r_user_id[294,Intercept]" "r_user_id[295,Intercept]" "r_user_id[296,Intercept]" "r_user_id[297,Intercept]"## [301] "r_user_id[298,Intercept]" "r_user_id[299,Intercept]" "r_user_id[300,Intercept]" "r_user_id[301,Intercept]" "r_user_id[302,Intercept]"## [306] "r_user_id[303,Intercept]" "r_user_id[304,Intercept]" "r_user_id[305,Intercept]" "r_user_id[306,Intercept]" "r_user_id[307,Intercept]"## [311] "r_user_id[308,Intercept]" "r_user_id[309,Intercept]" "r_user_id[310,Intercept]" "r_user_id[311,Intercept]" "r_user_id[312,Intercept]"## [316] "r_user_id[313,Intercept]" "r_user_id[314,Intercept]" "r_user_id[315,Intercept]" "r_user_id[316,Intercept]" "r_user_id[317,Intercept]"## [321] "r_user_id[318,Intercept]" "r_user_id[319,Intercept]" "r_user_id[320,Intercept]" "r_user_id[321,Intercept]" "r_user_id[322,Intercept]"## [326] "r_user_id[323,Intercept]" "r_user_id[324,Intercept]" "r_user_id[325,Intercept]" "r_user_id[326,Intercept]" "r_user_id[327,Intercept]"## [331] "r_user_id[328,Intercept]" "r_user_id[329,Intercept]" "r_user_id[330,Intercept]" "r_user_id[331,Intercept]" "r_user_id[332,Intercept]"## [336] "r_user_id[333,Intercept]" "r_user_id[334,Intercept]" "r_user_id[335,Intercept]" "r_user_id[336,Intercept]" "r_user_id[337,Intercept]"## [341] "r_user_id[338,Intercept]" "r_user_id[339,Intercept]" "r_user_id[340,Intercept]" "r_user_id[341,Intercept]" "r_user_id[342,Intercept]"## [346] "r_user_id[343,Intercept]" "r_user_id[344,Intercept]" "r_user_id[345,Intercept]" "r_user_id[346,Intercept]" "r_user_id[347,Intercept]"## [351] "r_user_id[348,Intercept]" "r_user_id[349,Intercept]" "r_user_id[350,Intercept]" "r_user_id[351,Intercept]" "r_user_id[352,Intercept]"## [356] "r_user_id[353,Intercept]" "r_user_id[354,Intercept]" "r_user_id[355,Intercept]" "r_user_id[356,Intercept]" "r_user_id[357,Intercept]"## [361] "r_user_id[358,Intercept]" "r_user_id[359,Intercept]" "r_user_id[360,Intercept]" "r_user_id[361,Intercept]" "r_user_id[362,Intercept]"## [366] "r_user_id[363,Intercept]" "r_user_id[364,Intercept]" "r_user_id[365,Intercept]" "r_user_id[366,Intercept]" "r_user_id[367,Intercept]"## [371] "r_user_id[368,Intercept]" "r_user_id[369,Intercept]" "r_user_id[370,Intercept]" "r_user_id[371,Intercept]" "r_user_id[372,Intercept]"## [376] "r_user_id[373,Intercept]" "r_user_id[374,Intercept]" "r_user_id[375,Intercept]" "r_user_id[376,Intercept]" "r_user_id[377,Intercept]"## [381] "r_user_id[378,Intercept]" "r_user_id[379,Intercept]" "r_user_id[380,Intercept]" "r_user_id[381,Intercept]" "r_user_id[382,Intercept]"## [386] "r_user_id[383,Intercept]" "r_user_id[384,Intercept]" "r_user_id[385,Intercept]" "r_user_id[386,Intercept]" "r_user_id[387,Intercept]"## [391] "r_user_id[388,Intercept]" "r_user_id[389,Intercept]" "r_user_id[390,Intercept]" "r_user_id[391,Intercept]" "r_user_id[392,Intercept]"## [396] "r_user_id[393,Intercept]" "r_user_id[394,Intercept]" "r_user_id[395,Intercept]" "r_user_id[396,Intercept]" "r_user_id[397,Intercept]"## [401] "r_user_id[398,Intercept]" "r_user_id[399,Intercept]" "r_user_id[400,Intercept]" "r_user_id[401,Intercept]" "r_user_id[402,Intercept]"## [406] "r_user_id[403,Intercept]" "r_user_id[404,Intercept]" "r_user_id[405,Intercept]" "r_user_id[406,Intercept]" "r_user_id[407,Intercept]"## [411] "r_user_id[408,Intercept]" "r_user_id[409,Intercept]" "r_user_id[410,Intercept]" "r_user_id[411,Intercept]" "r_user_id[412,Intercept]"## [416] "r_user_id[413,Intercept]" "r_user_id[414,Intercept]" "r_user_id[415,Intercept]" "r_user_id[416,Intercept]" "r_user_id[417,Intercept]"## [421] "r_user_id[418,Intercept]" "r_user_id[419,Intercept]" "r_user_id[420,Intercept]" "r_user_id[421,Intercept]" "r_user_id[422,Intercept]"## [426] "r_user_id[423,Intercept]" "r_user_id[424,Intercept]" "r_user_id[425,Intercept]" "r_user_id[426,Intercept]" "r_user_id[427,Intercept]"## [431] "r_user_id[428,Intercept]" "r_user_id[429,Intercept]" "r_user_id[430,Intercept]" "r_user_id[431,Intercept]" "r_user_id[432,Intercept]"## [436] "r_user_id[433,Intercept]" "r_user_id[434,Intercept]" "r_user_id[435,Intercept]" "r_user_id[436,Intercept]" "r_user_id[437,Intercept]"## [441] "r_user_id[438,Intercept]" "r_user_id[439,Intercept]" "r_user_id[440,Intercept]" "r_user_id[441,Intercept]" "r_user_id[442,Intercept]"## [446] "r_user_id[443,Intercept]" "r_user_id[444,Intercept]" "r_user_id[445,Intercept]" "r_user_id[446,Intercept]" "r_user_id[447,Intercept]"## [451] "r_user_id[448,Intercept]" "r_user_id[449,Intercept]" "r_user_id[450,Intercept]" "r_user_id[451,Intercept]" "r_user_id[452,Intercept]"## [456] "r_user_id[453,Intercept]" "r_user_id[454,Intercept]" "r_user_id[455,Intercept]" "r_user_id[456,Intercept]" "r_user_id[457,Intercept]"## [461] "r_user_id[458,Intercept]" "r_user_id[459,Intercept]" "r_user_id[460,Intercept]" "r_user_id[461,Intercept]" "r_user_id[462,Intercept]"## [466] "r_user_id[463,Intercept]" "r_user_id[464,Intercept]" "r_user_id[465,Intercept]" "r_user_id[466,Intercept]" "r_user_id[467,Intercept]"## [471] "r_user_id[468,Intercept]" "r_user_id[469,Intercept]" "r_user_id[470,Intercept]" "r_user_id[471,Intercept]" "r_user_id[472,Intercept]"## [476] "r_user_id[473,Intercept]" "r_user_id[474,Intercept]" "r_user_id[475,Intercept]" "r_user_id[476,Intercept]" "r_user_id[477,Intercept]"## [481] "r_user_id[478,Intercept]" "r_user_id[479,Intercept]" "r_user_id[480,Intercept]" "r_user_id[481,Intercept]" "r_user_id[482,Intercept]"## [486] "r_user_id[483,Intercept]" "r_user_id[484,Intercept]" "r_user_id[485,Intercept]" "r_user_id[486,Intercept]" "r_user_id[487,Intercept]"## [491] "r_user_id[488,Intercept]" "r_user_id[489,Intercept]" "r_user_id[490,Intercept]" "r_user_id[491,Intercept]" "r_user_id[492,Intercept]"## [496] "r_user_id[493,Intercept]" "r_user_id[494,Intercept]" "r_user_id[495,Intercept]" "r_user_id[496,Intercept]" "r_user_id[497,Intercept]"## [501] "r_user_id[498,Intercept]" "r_user_id[499,Intercept]" "r_user_id[500,Intercept]" "r_user_id[501,Intercept]" "r_user_id[502,Intercept]"## [506] "r_user_id[503,Intercept]" "r_user_id[504,Intercept]" "r_user_id[505,Intercept]" "r_user_id[506,Intercept]" "r_user_id[507,Intercept]"## [511] "r_user_id[508,Intercept]" "r_user_id[509,Intercept]" "r_user_id[510,Intercept]" "r_user_id[511,Intercept]" "r_user_id[512,Intercept]"## [516] "r_user_id[513,Intercept]" "r_user_id[514,Intercept]" "r_user_id[515,Intercept]" "r_user_id[516,Intercept]" "r_user_id[517,Intercept]"## [521] "r_user_id[518,Intercept]" "r_user_id[519,Intercept]" "r_user_id[520,Intercept]" "r_user_id[521,Intercept]" "r_user_id[522,Intercept]"## [526] "r_user_id[523,Intercept]" "r_user_id[524,Intercept]" "r_user_id[525,Intercept]" "r_user_id[526,Intercept]" "r_user_id[527,Intercept]"## [531] "r_user_id[528,Intercept]" "r_user_id[529,Intercept]" "r_user_id[530,Intercept]" "r_user_id[531,Intercept]" "r_user_id[532,Intercept]"## [536] "r_user_id[533,Intercept]" "r_user_id[534,Intercept]" "r_user_id[535,Intercept]" "r_user_id[536,Intercept]" "r_user_id[537,Intercept]"## [541] "r_user_id[538,Intercept]" "r_user_id[539,Intercept]" "r_user_id[540,Intercept]" "r_user_id[541,Intercept]" "r_user_id[542,Intercept]"## [546] "r_user_id[543,Intercept]" "r_user_id[544,Intercept]" "r_user_id[545,Intercept]" "r_user_id[546,Intercept]" "r_user_id[547,Intercept]"## [551] "r_user_id[548,Intercept]" "r_user_id[549,Intercept]" "r_user_id[550,Intercept]" "r_user_id[551,Intercept]" "r_user_id[552,Intercept]"## [556] "r_user_id[553,Intercept]" "r_user_id[554,Intercept]" "r_user_id[555,Intercept]" "r_user_id[556,Intercept]" "r_user_id[557,Intercept]"## [561] "r_user_id[558,Intercept]" "r_user_id[559,Intercept]" "r_user_id[560,Intercept]" "r_user_id[561,Intercept]" "r_user_id[562,Intercept]"## [566] "r_user_id[563,Intercept]" "r_user_id[564,Intercept]" "r_user_id[565,Intercept]" "r_user_id[566,Intercept]" "r_user_id[567,Intercept]"## [571] "r_user_id[568,Intercept]" "r_user_id[569,Intercept]" "r_user_id[570,Intercept]" "r_user_id[571,Intercept]" "r_user_id[572,Intercept]"## [576] "r_user_id[573,Intercept]" "r_user_id[574,Intercept]" "r_user_id[575,Intercept]" "r_user_id[576,Intercept]" "r_user_id[577,Intercept]"## [581] "r_user_id[578,Intercept]" "r_user_id[579,Intercept]" "r_user_id[580,Intercept]" "r_user_id[581,Intercept]" "r_user_id[582,Intercept]"## [586] "r_user_id[583,Intercept]" "r_user_id[584,Intercept]" "r_user_id[585,Intercept]" "r_user_id[586,Intercept]" "r_user_id[587,Intercept]"## [591] "r_user_id[588,Intercept]" "r_user_id[589,Intercept]" "r_user_id[590,Intercept]" "r_user_id[591,Intercept]" "r_user_id[592,Intercept]"## [596] "r_user_id[593,Intercept]" "r_user_id[594,Intercept]" "r_user_id[595,Intercept]" "r_user_id[596,Intercept]" "r_user_id[597,Intercept]"## [601] "r_user_id[598,Intercept]" "r_user_id[599,Intercept]" "r_user_id[600,Intercept]" "r_user_id[601,Intercept]" "r_user_id[602,Intercept]"## [606] "r_user_id[603,Intercept]" "r_user_id[604,Intercept]" "r_user_id[605,Intercept]" "r_user_id[606,Intercept]" "r_user_id[607,Intercept]"## [611] "r_user_id[608,Intercept]" "r_user_id[609,Intercept]" "r_user_id[610,Intercept]" "r_user_id[611,Intercept]" "r_user_id[612,Intercept]"## [616] "r_user_id[613,Intercept]" "r_user_id[614,Intercept]" "r_user_id[615,Intercept]" "r_user_id[616,Intercept]" "r_user_id[617,Intercept]"## [621] "r_user_id[618,Intercept]" "r_user_id[619,Intercept]" "r_user_id[620,Intercept]" "r_user_id[621,Intercept]" "r_user_id[622,Intercept]"## [626] "r_user_id[623,Intercept]" "r_user_id[624,Intercept]" "r_user_id[625,Intercept]" "r_user_id[626,Intercept]" "r_user_id[627,Intercept]"## [631] "r_user_id[628,Intercept]" "r_user_id[629,Intercept]" "r_user_id[630,Intercept]" "r_user_id[631,Intercept]" "r_user_id[632,Intercept]"## [636] "r_user_id[633,Intercept]" "r_user_id[634,Intercept]" "r_user_id[635,Intercept]" "r_user_id[636,Intercept]" "r_user_id[637,Intercept]"## [641] "r_user_id[638,Intercept]" "r_user_id[639,Intercept]" "r_user_id[640,Intercept]" "r_user_id[641,Intercept]" "r_user_id[642,Intercept]"## [646] "r_user_id[643,Intercept]" "r_user_id[644,Intercept]" "r_user_id[645,Intercept]" "r_user_id[646,Intercept]" "r_user_id[647,Intercept]"## [651] "r_user_id[648,Intercept]" "r_user_id[649,Intercept]" "r_user_id[650,Intercept]" "r_user_id[651,Intercept]" "r_user_id[652,Intercept]"## [656] "r_user_id[653,Intercept]" "r_user_id[654,Intercept]" "r_user_id[655,Intercept]" "r_user_id[656,Intercept]" "r_user_id[657,Intercept]"## [661] "r_user_id[658,Intercept]" "r_user_id[659,Intercept]" "r_user_id[660,Intercept]" "r_user_id[661,Intercept]" "r_user_id[662,Intercept]"## [666] "r_user_id[663,Intercept]" "r_user_id[664,Intercept]" "r_user_id[665,Intercept]" "r_user_id[666,Intercept]" "r_user_id[667,Intercept]"## [671] "r_user_id[668,Intercept]" "r_user_id[669,Intercept]" "r_user_id[670,Intercept]" "r_user_id[671,Intercept]" "r_user_id[672,Intercept]"## [676] "r_user_id[673,Intercept]" "r_user_id[674,Intercept]" "r_user_id[675,Intercept]" "r_user_id[676,Intercept]" "r_user_id[677,Intercept]"## [681] "r_user_id[678,Intercept]" "r_user_id[679,Intercept]" "r_user_id[680,Intercept]" "r_user_id[681,Intercept]" "r_user_id[682,Intercept]"## [686] "r_user_id[683,Intercept]" "r_user_id[684,Intercept]" "r_user_id[685,Intercept]" "r_user_id[686,Intercept]" "r_user_id[687,Intercept]"## [691] "r_user_id[688,Intercept]" "r_user_id[689,Intercept]" "r_user_id[690,Intercept]" "r_user_id[691,Intercept]" "r_user_id[692,Intercept]"## [696] "r_user_id[693,Intercept]" "r_user_id[694,Intercept]" "r_user_id[695,Intercept]" "r_user_id[696,Intercept]" "r_user_id[697,Intercept]"## [701] "r_user_id[698,Intercept]" "r_user_id[699,Intercept]" "r_user_id[700,Intercept]" "r_user_id[701,Intercept]" "r_user_id[702,Intercept]"## [706] "r_user_id[703,Intercept]" "r_user_id[704,Intercept]" "r_user_id[705,Intercept]" "r_user_id[706,Intercept]" "r_user_id[707,Intercept]"## [711] "r_user_id[708,Intercept]" "r_user_id[709,Intercept]" "r_user_id[710,Intercept]" "r_user_id[711,Intercept]" "r_user_id[712,Intercept]"## [716] "r_user_id[713,Intercept]" "r_user_id[714,Intercept]" "r_user_id[715,Intercept]" "r_user_id[716,Intercept]" "r_user_id[717,Intercept]"## [721] "r_user_id[718,Intercept]" "r_user_id[719,Intercept]" "r_user_id[720,Intercept]" "r_user_id[721,Intercept]" "r_user_id[722,Intercept]"## [726] "r_user_id[723,Intercept]" "r_user_id[724,Intercept]" "r_user_id[725,Intercept]" "r_user_id[726,Intercept]" "r_user_id[727,Intercept]"## [731] "r_user_id[728,Intercept]" "r_user_id[729,Intercept]" "r_user_id[730,Intercept]" "r_user_id[731,Intercept]" "r_user_id[732,Intercept]"## [736] "r_user_id[733,Intercept]" "r_user_id[734,Intercept]" "r_user_id[735,Intercept]" "r_user_id[736,Intercept]" "r_user_id[737,Intercept]"## [741] "r_user_id[738,Intercept]" "r_user_id[739,Intercept]" "r_user_id[740,Intercept]" "r_user_id[741,Intercept]" "r_user_id[742,Intercept]"## [746] "r_user_id[743,Intercept]" "r_user_id[744,Intercept]" "r_user_id[745,Intercept]" "r_user_id[746,Intercept]" "r_user_id[747,Intercept]"## [751] "r_user_id[748,Intercept]" "r_user_id[749,Intercept]" "r_user_id[750,Intercept]" "r_user_id[751,Intercept]" "r_user_id[752,Intercept]"## [756] "r_user_id[753,Intercept]" "r_user_id[754,Intercept]" "r_user_id[755,Intercept]" "r_user_id[756,Intercept]" "r_user_id[757,Intercept]"## [761] "r_user_id[758,Intercept]" "r_user_id[759,Intercept]" "r_user_id[760,Intercept]" "r_user_id[761,Intercept]" "r_user_id[762,Intercept]"## [766] "r_user_id[763,Intercept]" "r_user_id[764,Intercept]" "r_user_id[765,Intercept]" "r_user_id[766,Intercept]" "r_user_id[767,Intercept]"## [771] "r_user_id[768,Intercept]" "r_user_id[769,Intercept]" "r_user_id[770,Intercept]" "r_user_id[771,Intercept]" "r_user_id[772,Intercept]"## [776] "r_user_id[773,Intercept]" "r_user_id[774,Intercept]" "r_user_id[775,Intercept]" "r_user_id[776,Intercept]" "r_user_id[777,Intercept]"## [781] "r_user_id[778,Intercept]" "r_user_id[779,Intercept]" "r_user_id[780,Intercept]" "r_user_id[781,Intercept]" "r_user_id[782,Intercept]"## [786] "r_user_id[783,Intercept]" "r_user_id[784,Intercept]" "r_user_id[785,Intercept]" "r_user_id[786,Intercept]" "r_user_id[787,Intercept]"## [791] "r_user_id[788,Intercept]" "r_user_id[789,Intercept]" "r_user_id[790,Intercept]" "r_user_id[791,Intercept]" "r_user_id[792,Intercept]"## [796] "r_user_id[793,Intercept]" "r_user_id[794,Intercept]" "r_user_id[795,Intercept]" "r_user_id[796,Intercept]" "r_user_id[797,Intercept]"## [801] "r_user_id[798,Intercept]" "r_user_id[799,Intercept]" "r_user_id[800,Intercept]" "r_user_id[801,Intercept]" "r_user_id[802,Intercept]"## [806] "r_user_id[803,Intercept]" "r_user_id[804,Intercept]" "r_user_id[805,Intercept]" "r_user_id[806,Intercept]" "r_user_id[807,Intercept]"## [811] "r_user_id[808,Intercept]" "r_user_id[809,Intercept]" "r_user_id[810,Intercept]" "r_user_id[811,Intercept]" "r_user_id[812,Intercept]"## [816] "r_user_id[813,Intercept]" "r_user_id[814,Intercept]" "r_user_id[815,Intercept]" "r_user_id[816,Intercept]" "r_user_id[817,Intercept]"## [821] "r_user_id[818,Intercept]" "r_user_id[819,Intercept]" "r_user_id[820,Intercept]" "r_user_id[821,Intercept]" "r_user_id[822,Intercept]"## [826] "r_user_id[823,Intercept]" "r_user_id[824,Intercept]" "r_user_id[825,Intercept]" "r_user_id[826,Intercept]" "r_user_id[827,Intercept]"## [831] "r_user_id[828,Intercept]" "r_user_id[829,Intercept]" "r_user_id[830,Intercept]" "r_user_id[831,Intercept]" "r_user_id[832,Intercept]"## [836] "r_user_id[833,Intercept]" "r_user_id[834,Intercept]" "r_user_id[835,Intercept]" "r_user_id[836,Intercept]" "r_user_id[837,Intercept]"## [841] "r_user_id[838,Intercept]" "r_user_id[839,Intercept]" "r_user_id[840,Intercept]" "r_user_id[841,Intercept]" "r_user_id[842,Intercept]"## [846] "r_user_id[843,Intercept]" "r_user_id[844,Intercept]" "r_user_id[845,Intercept]" "r_user_id[846,Intercept]" "r_user_id[847,Intercept]"## [851] "r_user_id[848,Intercept]" "r_user_id[849,Intercept]" "r_user_id[850,Intercept]" "r_user_id[851,Intercept]" "r_user_id[852,Intercept]"## [856] "r_user_id[853,Intercept]" "r_user_id[854,Intercept]" "r_user_id[855,Intercept]" "r_user_id[856,Intercept]" "r_user_id[857,Intercept]"## [861] "r_user_id[858,Intercept]" "r_user_id[859,Intercept]" "r_user_id[860,Intercept]" "r_user_id[861,Intercept]" "r_user_id[862,Intercept]"## [866] "r_user_id[863,Intercept]" "r_user_id[864,Intercept]" "r_user_id[865,Intercept]" "r_user_id[866,Intercept]" "r_user_id[867,Intercept]"## [871] "r_user_id[868,Intercept]" "r_user_id[869,Intercept]" "r_user_id[870,Intercept]" "r_user_id[871,Intercept]" "r_user_id[872,Intercept]"## [876] "r_user_id[873,Intercept]" "r_user_id[874,Intercept]" "r_user_id[875,Intercept]" "r_user_id[876,Intercept]" "r_user_id[877,Intercept]"## [881] "r_user_id[878,Intercept]" "r_user_id[879,Intercept]" "r_user_id[880,Intercept]" "r_user_id[881,Intercept]" "r_user_id[882,Intercept]"## [886] "r_user_id[883,Intercept]" "r_user_id[884,Intercept]" "r_user_id[885,Intercept]" "r_user_id[886,Intercept]" "r_user_id[887,Intercept]"## [891] "r_user_id[888,Intercept]" "r_user_id[889,Intercept]" "r_user_id[890,Intercept]" "r_user_id[891,Intercept]" "r_user_id[892,Intercept]"## [896] "r_user_id[893,Intercept]" "r_user_id[894,Intercept]" "r_user_id[895,Intercept]" "r_user_id[896,Intercept]" "r_user_id[897,Intercept]"## [901] "r_user_id[898,Intercept]" "r_user_id[899,Intercept]" "r_user_id[900,Intercept]" "r_user_id[901,Intercept]" "r_user_id[902,Intercept]"## [906] "r_user_id[903,Intercept]" "r_user_id[904,Intercept]" "r_user_id[905,Intercept]" "r_user_id[906,Intercept]" "r_user_id[907,Intercept]"## [911] "r_user_id[908,Intercept]" "r_user_id[909,Intercept]" "r_user_id[910,Intercept]" "r_user_id[911,Intercept]" "r_user_id[912,Intercept]"## [916] "r_user_id[913,Intercept]" "r_user_id[914,Intercept]" "r_user_id[915,Intercept]" "r_user_id[916,Intercept]" "r_user_id[917,Intercept]"## [921] "r_user_id[918,Intercept]" "r_user_id[919,Intercept]" "r_user_id[920,Intercept]" "r_user_id[921,Intercept]" "r_user_id[922,Intercept]"## [926] "r_user_id[923,Intercept]" "r_user_id[924,Intercept]" "r_user_id[925,Intercept]" "r_user_id[926,Intercept]" "r_user_id[927,Intercept]"## [931] "r_user_id[928,Intercept]" "r_user_id[929,Intercept]" "r_user_id[930,Intercept]" "r_user_id[931,Intercept]" "r_user_id[932,Intercept]"## [936] "r_user_id[933,Intercept]" "r_user_id[934,Intercept]" "r_user_id[935,Intercept]" "r_user_id[936,Intercept]" "r_user_id[937,Intercept]"## [941] "r_user_id[938,Intercept]" "r_user_id[939,Intercept]" "r_user_id[940,Intercept]" "r_user_id[941,Intercept]" "r_user_id[942,Intercept]"## [946] "r_user_id[943,Intercept]" "r_user_id[944,Intercept]" "r_user_id[945,Intercept]" "r_user_id[946,Intercept]" "r_user_id[947,Intercept]"## [951] "r_user_id[948,Intercept]" "r_user_id[949,Intercept]" "r_user_id[950,Intercept]" "r_user_id[951,Intercept]" "r_user_id[952,Intercept]"## [956] "r_user_id[953,Intercept]" "r_user_id[954,Intercept]" "r_user_id[955,Intercept]" "r_user_id[956,Intercept]" "r_user_id[957,Intercept]"## [961] "r_user_id[958,Intercept]" "r_user_id[959,Intercept]" "r_user_id[960,Intercept]" "r_user_id[961,Intercept]" "r_user_id[962,Intercept]"## [966] "r_user_id[963,Intercept]" "r_user_id[964,Intercept]" "r_user_id[965,Intercept]" "r_user_id[966,Intercept]" "r_user_id[967,Intercept]"## [971] "r_user_id[968,Intercept]" "r_user_id[969,Intercept]" "r_user_id[970,Intercept]" "r_user_id[971,Intercept]" "r_user_id[972,Intercept]"## [976] "r_user_id[973,Intercept]" "r_user_id[974,Intercept]" "r_user_id[975,Intercept]" "r_user_id[976,Intercept]" "r_user_id[977,Intercept]"## [981] "r_user_id[978,Intercept]" "r_user_id[979,Intercept]" "r_user_id[980,Intercept]" "r_user_id[981,Intercept]" "r_user_id[982,Intercept]"## [986] "r_user_id[983,Intercept]" "r_user_id[984,Intercept]" "r_user_id[985,Intercept]" "r_user_id[986,Intercept]" "r_user_id[987,Intercept]"## [991] "r_user_id[988,Intercept]" "r_user_id[989,Intercept]" "r_user_id[990,Intercept]" "r_user_id[991,Intercept]" "r_user_id[992,Intercept]"## [996] "r_user_id[993,Intercept]" "r_user_id[994,Intercept]" "r_user_id[995,Intercept]" "r_user_id[996,Intercept]" "r_user_id[997,Intercept]"## [ reached getOption("max.print") -- omitted 17918 entries ]Pull random vars
The random effect name is
r_user_idWe use brackets to assign new names
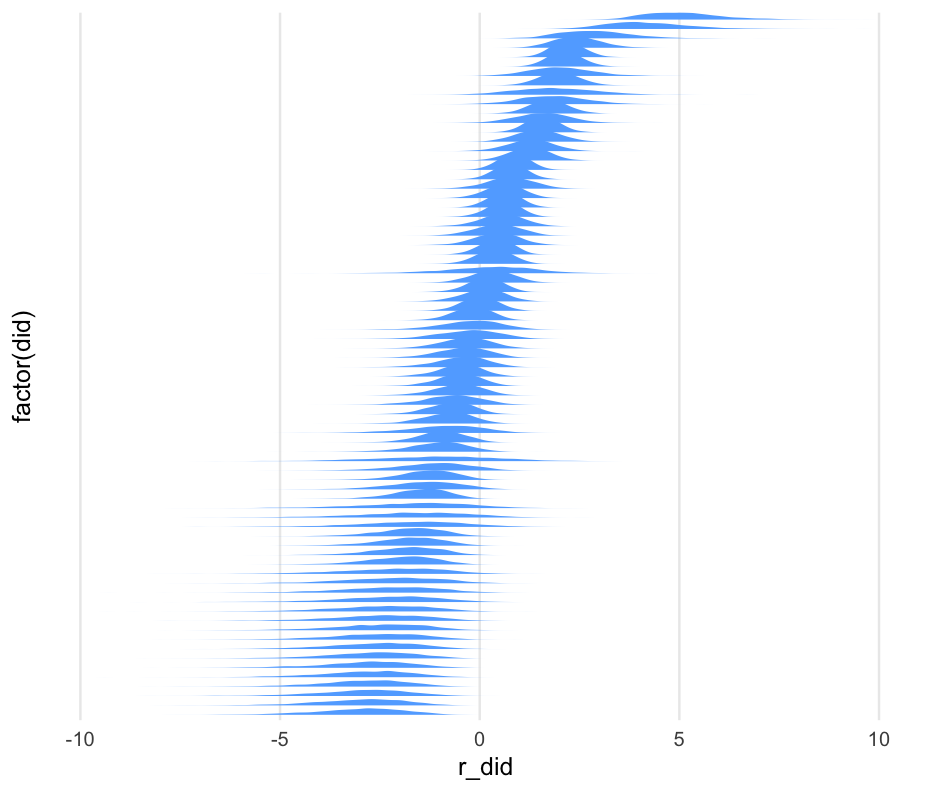
m0_id_re <- gather_draws(m0_b, r_user_id[id, term])m0_id_re## # A tibble: 37,816,000 x 7## # Groups: id, term, .variable [9,454]## id term .chain .iteration .draw .variable .value## <int> <chr> <int> <int> <int> <chr> <dbl>## 1 1 Intercept 1 1 1 r_user_id -0.0826422## 2 1 Intercept 1 2 2 r_user_id -0.567424 ## 3 1 Intercept 1 3 3 r_user_id -0.546465 ## 4 1 Intercept 1 4 4 r_user_id -1.88725 ## 5 1 Intercept 1 5 5 r_user_id 1.12419 ## 6 1 Intercept 1 6 6 r_user_id -2.31118 ## 7 1 Intercept 1 7 7 r_user_id -0.249097 ## 8 1 Intercept 1 8 8 r_user_id -3.61696 ## 9 1 Intercept 1 9 9 r_user_id -1.6264 ## 10 1 Intercept 1 10 10 r_user_id 0.158484 ## # … with 37,815,990 more rowsCompute credible intervals
I recognize this part is complex, but you'll have the code for reference
id_qtiles <- m0_id_re %>% group_by(id) %>% summarize( probs = c("median", "lower", "upper"), qtiles = quantile(.value,probs = c(0.5, 0.025, 0.975)) ) %>% ungroup() id_qtiles## # A tibble: 28,362 x 3## id probs qtiles## <int> <chr> <dbl>## 1 1 median -0.6739 ## 2 1 lower -3.31484 ## 3 1 upper 1.746887 ## 4 2 median 0.9492155## 5 2 lower -1.605349 ## 6 2 upper 3.453275 ## 7 3 median -0.648632 ## 8 3 lower -3.341358 ## 9 3 upper 1.803793 ## 10 4 median 0.9214355## # … with 28,352 more rowsMove it wider
id_qtiles <- id_qtiles %>% pivot_wider(names_from = "probs", values_from = "qtiles") %>% mutate(id = fct_reorder(factor(id), median))id_qtiles## # A tibble: 9,454 x 4## id median lower upper## <fct> <dbl> <dbl> <dbl>## 1 1 -0.6739 -3.31484 1.746887## 2 2 0.9492155 -1.605349 3.453275## 3 3 -0.648632 -3.341358 1.803793## 4 4 0.9214355 -1.517492 3.542017## 5 5 -0.626921 -3.264422 1.725803## 6 6 0.930557 -1.599318 3.539588## 7 7 -0.6482415 -3.349034 1.817699## 8 8 -0.6443055 -3.212276 1.697759## 9 9 -0.6500125 -3.371763 1.892287## 10 10 -1.029665 -3.648958 1.221002## # … with 9,444 more rowsm1_b <- brm(is_positive_sentiment ~ trump_in_description + has_antifa_hashtag + log(favorite_count + 1) + (1|user_id), data = blm, family = bernoulli(link = "logit"), cores = 4, backend = "cmdstanr")## -\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\Running MCMC with 4 parallel chains...## ## Chain 3 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 4 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 1 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 2 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 3 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 1 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 2 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 4 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 3 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 1 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 2 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 4 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 3 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 1 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 2 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 4 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 3 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 1 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 2 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 4 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 3 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 2 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 1 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 4 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 3 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 2 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 1 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 4 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 3 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 2 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 1 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 4 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 3 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 2 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 4 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 1 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 3 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 2 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 1 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 4 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 3 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 3 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 2 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 2 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 1 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 1 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 4 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 4 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 3 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 2 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 1 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 4 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 3 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 2 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 1 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 4 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 3 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 2 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 1 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 4 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 3 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 2 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 1 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 4 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 3 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 2 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 1 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 4 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 3 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 2 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 1 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 4 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 3 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 2 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 1 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 4 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 3 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 2 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 1 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 4 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 3 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 2 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 1 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 4 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 3 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 3 finished in 125.6 seconds.## Chain 2 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 2 finished in 127.9 seconds.## Chain 1 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 4 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 1 finished in 128.1 seconds.## Chain 4 finished in 128.1 seconds.## ## All 4 chains finished successfully.## Mean chain execution time: 127.4 seconds.## Total execution time: 128.6 seconds.Summary
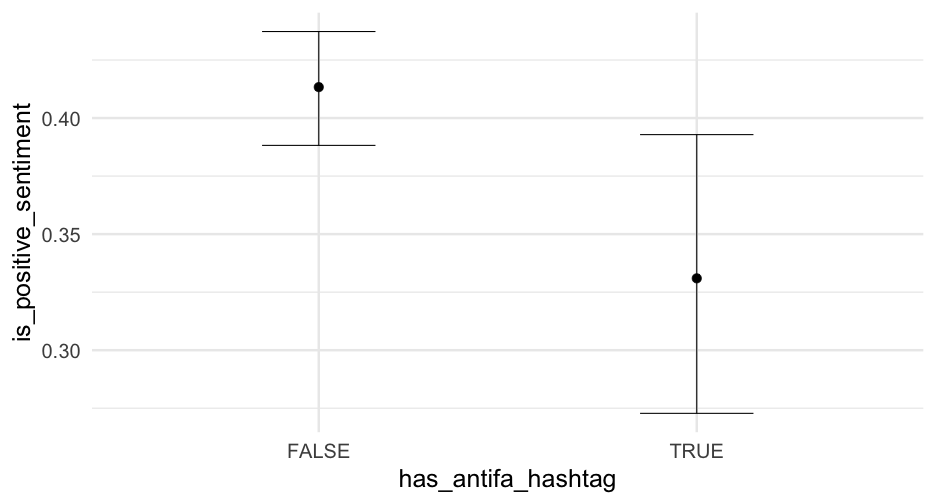
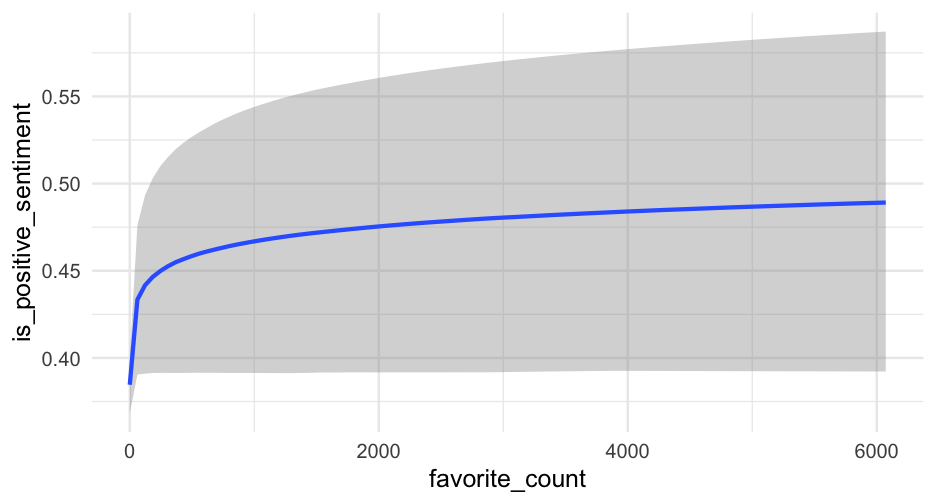
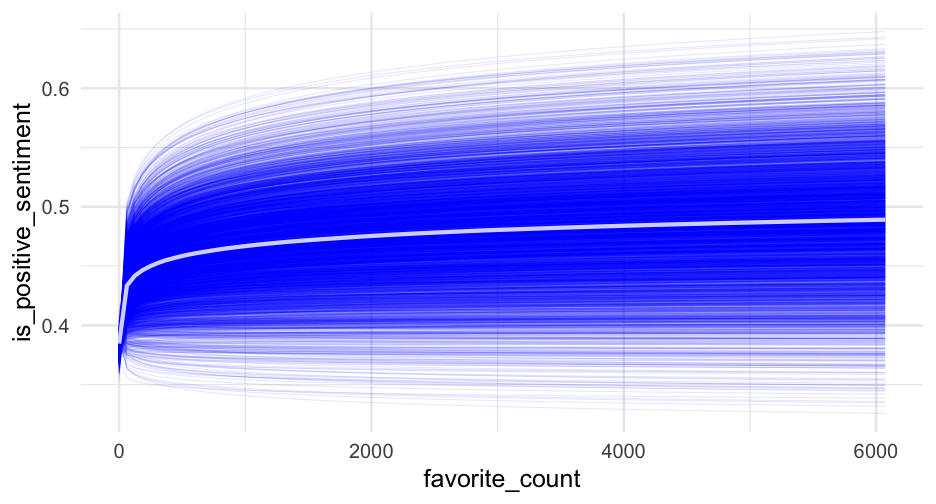
summary(m1_b)## Family: bernoulli ## Links: mu = logit ## Formula: is_positive_sentiment ~ trump_in_description + has_antifa_hashtag + log(favorite_count + 1) + (1 | user_id) ## Data: blm (Number of observations: 14339) ## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;## total post-warmup samples = 4000## ## Group-Level Effects: ## ~user_id (Number of levels: 9454) ## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS## sd(Intercept) 1.51 0.07 1.37 1.65 1.01 663 1341## ## Population-Level Effects: ## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS## Intercept -0.47 0.03 -0.54 -0.40 1.00 2715 3062## trump_in_descriptionTRUE -0.36 0.19 -0.74 0.01 1.00 2735 2912## has_antifa_hashtagTRUE -0.35 0.13 -0.61 -0.10 1.00 3810 2924## logfavorite_countP1 0.05 0.03 -0.00 0.10 1.00 3507 3361## ## Samples were drawn using sample(hmc). For each parameter, Bulk_ESS## and Tail_ESS are effective sample size measures, and Rhat is the potential## scale reduction factor on split chains (at convergence, Rhat = 1).Posteriors
What's the likelihood that our posterior mean for the log of favorite counts is positive?
m1_posterior <- get_parameters(m1_b)sum(m1_posterior$b_logfavorite_countP1 > 0) / nrow(m1_posterior)## [1] 0.9717597% probability! But note - this would probably just miss "significance", with a frequentist approach because of the use of two-tailed tests.
Warnings
As I was playing around with different models, I sometimes ran into warnings.
These were easy to solve in this case by just log-transforming the predictor variables that were highly skewed.
For general guidance, see here.
New data
Lung cancer data: Patients nested in doctors
hdp <- read_csv("https://stats.idre.ucla.edu/stat/data/hdp.csv") %>% janitor::clean_names() %>% select(did, tumorsize, pain, lungcapacity, age, remission)hdp## # A tibble: 8,525 x 6## did tumorsize pain lungcapacity age remission## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 67.98120 4 0.8010882 64.96824 0## 2 1 64.70246 2 0.3264440 53.91714 0## 3 1 51.56700 6 0.5650309 53.34730 0## 4 1 86.43799 3 0.8484109 41.36804 0## 5 1 53.40018 3 0.8864910 46.80042 0## 6 1 51.65727 4 0.7010307 51.92936 0## 7 1 78.91707 3 0.8908539 53.82926 0## 8 1 69.83325 3 0.6608795 46.56223 0## 9 1 62.85259 4 0.9088714 54.38936 0## 10 1 71.77790 5 0.9593268 50.54465 0## # … with 8,515 more rowsPredict remission
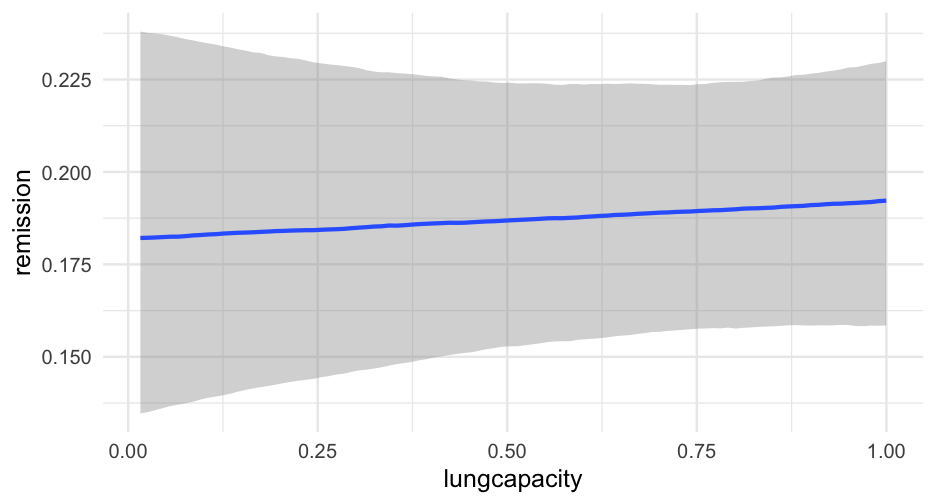
Build a model where age, lung capacity, and tumor size predict whether or not the patient was in remission.
Build the model so you can evaluate whether or not the relation between the tumor size and likelihood of remission depends on age
Allow the intercept to vary by the doctor ID.
Fit the model using brms
05:00
Lung cancer remission model
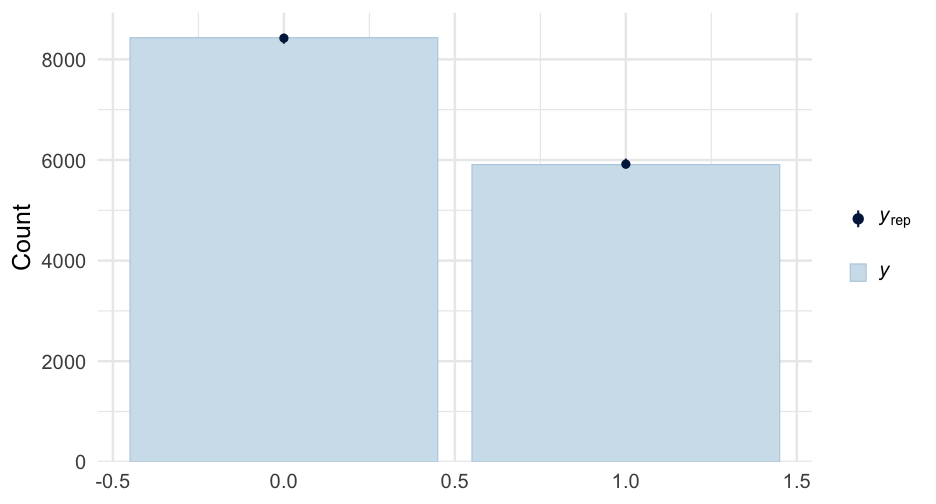
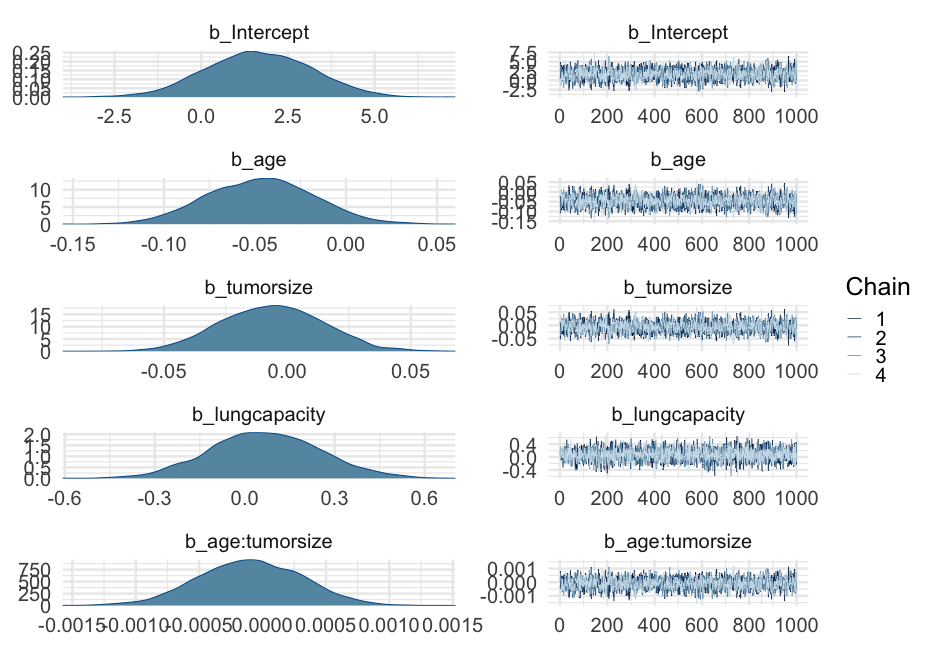
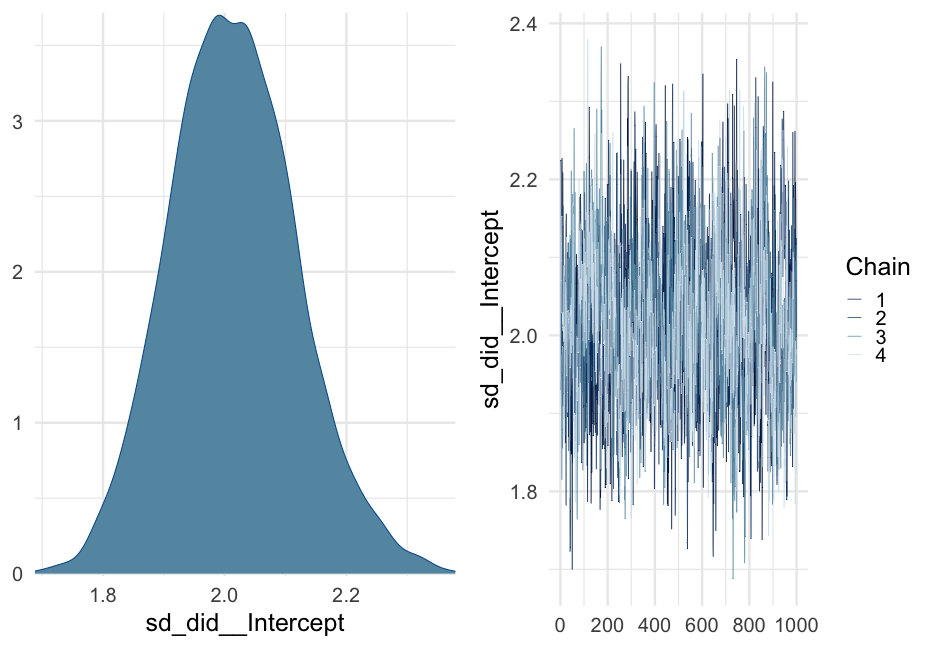
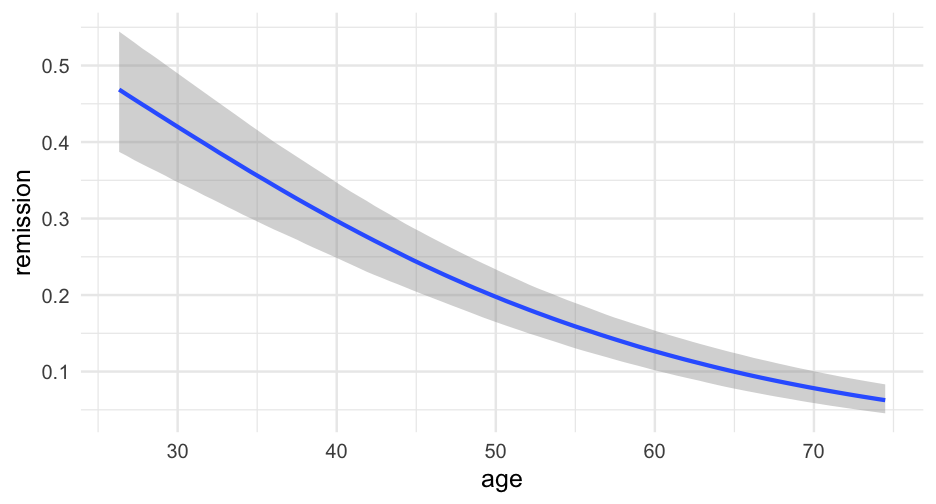
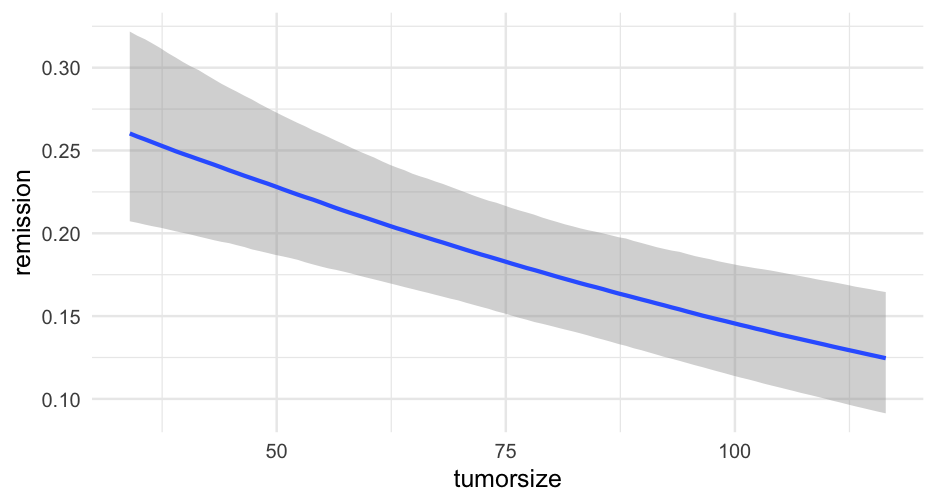
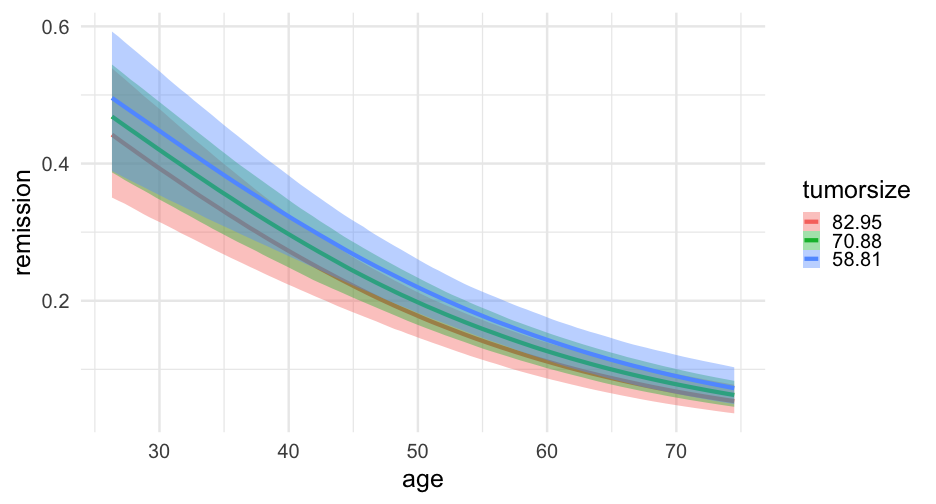
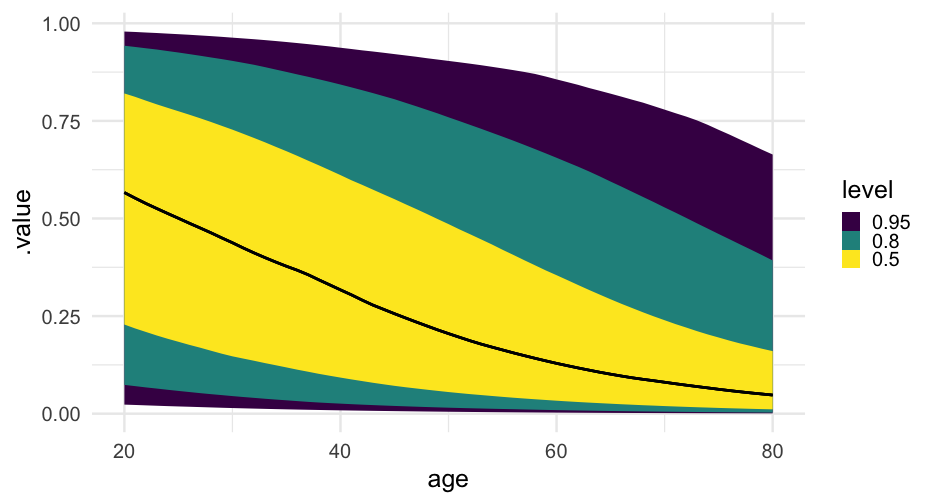
lc <- brm( remission ~ age * tumorsize + lungcapacity + (1|did), data = hdp, family = bernoulli(link = "logit"), cores = 4, backend = "cmdstan")## -\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\|/-\Running MCMC with 4 parallel chains...## ## Chain 1 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 2 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 3 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 4 Iteration: 1 / 2000 [ 0%] (Warmup) ## Chain 3 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 1 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 4 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 2 Iteration: 100 / 2000 [ 5%] (Warmup) ## Chain 3 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 1 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 2 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 4 Iteration: 200 / 2000 [ 10%] (Warmup) ## Chain 3 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 1 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 2 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 4 Iteration: 300 / 2000 [ 15%] (Warmup) ## Chain 3 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 2 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 1 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 4 Iteration: 400 / 2000 [ 20%] (Warmup) ## Chain 2 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 3 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 1 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 4 Iteration: 500 / 2000 [ 25%] (Warmup) ## Chain 2 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 3 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 1 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 4 Iteration: 600 / 2000 [ 30%] (Warmup) ## Chain 2 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 3 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 1 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 4 Iteration: 700 / 2000 [ 35%] (Warmup) ## Chain 3 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 2 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 1 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 4 Iteration: 800 / 2000 [ 40%] (Warmup) ## Chain 2 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 3 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 1 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 4 Iteration: 900 / 2000 [ 45%] (Warmup) ## Chain 2 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 2 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 3 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 3 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 1 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 1 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 4 Iteration: 1000 / 2000 [ 50%] (Warmup) ## Chain 4 Iteration: 1001 / 2000 [ 50%] (Sampling) ## Chain 3 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 1 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 2 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 4 Iteration: 1100 / 2000 [ 55%] (Sampling) ## Chain 3 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 1 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 4 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 2 Iteration: 1200 / 2000 [ 60%] (Sampling) ## Chain 3 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 1 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 4 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 2 Iteration: 1300 / 2000 [ 65%] (Sampling) ## Chain 3 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 1 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 4 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 2 Iteration: 1400 / 2000 [ 70%] (Sampling) ## Chain 3 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 1 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 4 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 1 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 3 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 2 Iteration: 1500 / 2000 [ 75%] (Sampling) ## Chain 4 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 1 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 3 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 4 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 2 Iteration: 1600 / 2000 [ 80%] (Sampling) ## Chain 1 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 3 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 4 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 2 Iteration: 1700 / 2000 [ 85%] (Sampling) ## Chain 1 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 3 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 4 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 2 Iteration: 1800 / 2000 [ 90%] (Sampling) ## Chain 1 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 1 finished in 302.2 seconds.## Chain 3 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 3 finished in 304.3 seconds.## Chain 4 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 4 finished in 305.6 seconds.## Chain 2 Iteration: 1900 / 2000 [ 95%] (Sampling) ## Chain 2 Iteration: 2000 / 2000 [100%] (Sampling) ## Chain 2 finished in 321.1 seconds.## ## All 4 chains finished successfully.## Mean chain execution time: 308.3 seconds.## Total execution time: 321.6 seconds.Model summary
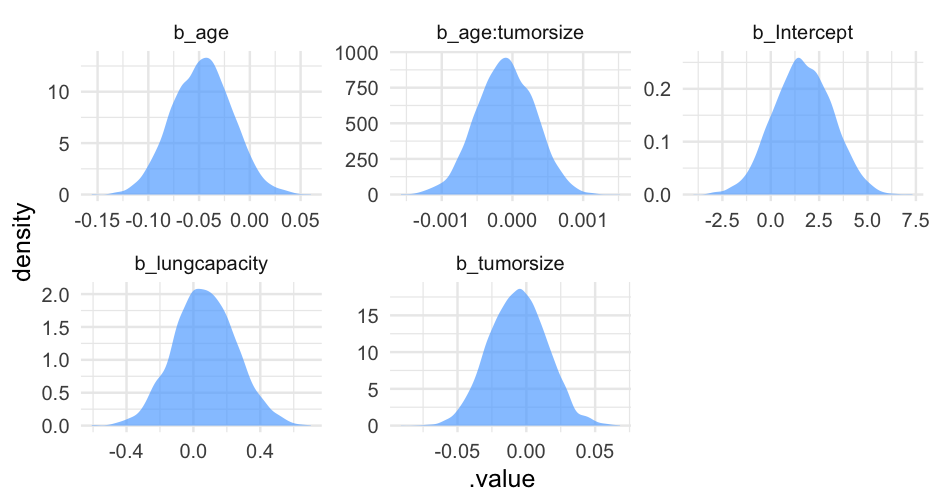
summary(lc)## Family: bernoulli ## Links: mu = logit ## Formula: remission ~ age * tumorsize + lungcapacity + (1 | did) ## Data: hdp (Number of observations: 8525) ## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;## total post-warmup samples = 4000## ## Group-Level Effects: ## ~did (Number of levels: 407) ## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS## sd(Intercept) 2.01 0.10 1.82 2.23 1.00 599 1603## ## Population-Level Effects: ## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS## Intercept 1.66 1.51 -1.31 4.57 1.00 1925 2782## age -0.05 0.03 -0.10 0.01 1.00 1919 2538## tumorsize -0.01 0.02 -0.05 0.03 1.00 1931 2833## lungcapacity 0.07 0.19 -0.29 0.44 1.00 4872 3061## age:tumorsize -0.00 0.00 -0.00 0.00 1.00 1905 2810## ## Samples were drawn using sample(hmc). For each parameter, Bulk_ESS## and Tail_ESS are effective sample size measures, and Rhat is the potential## scale reduction factor on split chains (at convergence, Rhat = 1).pred_tumor## # A tibble: 555,100 x 9## # Groups: age, lungcapacity, tumorsize, did, .row [5,551]## age lungcapacity tumorsize did .row .chain .iteration .draw .value## <int> <dbl> <int> <dbl> <int> <int> <int> <int> <dbl>## 1 20 0.7740865 30 -999 1 NA NA 22 0.8898250## 2 20 0.7740865 30 -999 1 NA NA 167 0.1359217## 3 20 0.7740865 30 -999 1 NA NA 296 0.4548241## 4 20 0.7740865 30 -999 1 NA NA 327 0.9331879## 5 20 0.7740865 30 -999 1 NA NA 371 0.3171560## 6 20 0.7740865 30 -999 1 NA NA 392 0.9858511## 7 20 0.7740865 30 -999 1 NA NA 446 0.9968882## 8 20 0.7740865 30 -999 1 NA NA 461 0.3545641## 9 20 0.7740865 30 -999 1 NA NA 555 0.8710298## 10 20 0.7740865 30 -999 1 NA NA 559 0.7410803## # … with 555,090 more rowsVariance by Doctor
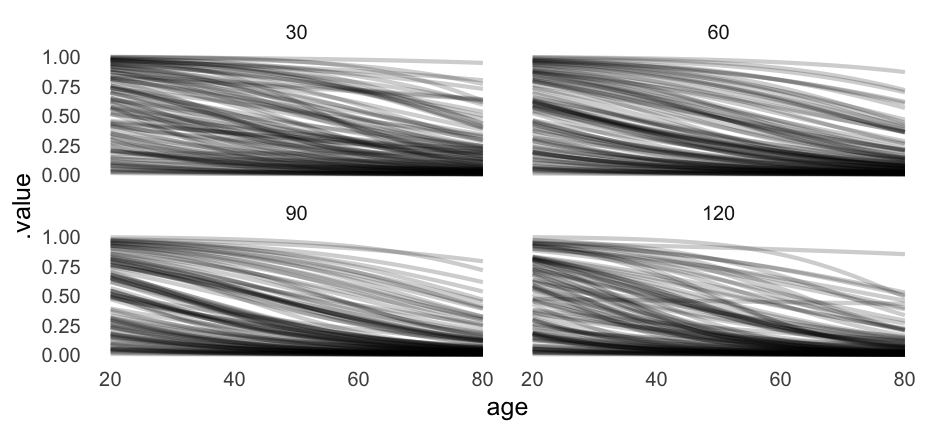
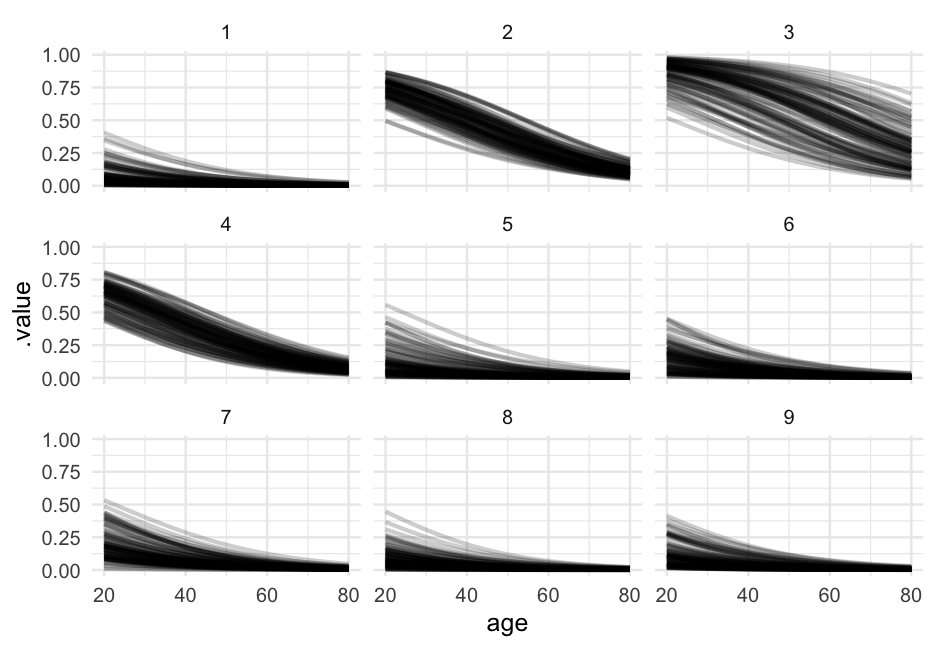
Let's look at the relation between age and proability of remission for each of the first nine doctors.
pred_age_doctor <- expand.grid( did = unique(hdp$did)[1:9], age = 20:80, tumorsize = mean(hdp$tumorsize), lungcapacity = mean(hdp$lungcapacity) ) %>% add_fitted_draws(model = lc, n = 100)pred_age_doctor## # A tibble: 54,900 x 9## # Groups: did, age, tumorsize, lungcapacity, .row [549]## did age tumorsize lungcapacity .row .chain .iteration .draw .value## <dbl> <int> <dbl> <dbl> <int> <int> <int> <int> <dbl>## 1 1 20 70.88067 0.7740865 1 NA NA 38 0.3467106 ## 2 1 20 70.88067 0.7740865 1 NA NA 73 0.1107260 ## 3 1 20 70.88067 0.7740865 1 NA NA 99 0.005263299## 4 1 20 70.88067 0.7740865 1 NA NA 107 0.02089231 ## 5 1 20 70.88067 0.7740865 1 NA NA 217 0.1016657 ## 6 1 20 70.88067 0.7740865 1 NA NA 241 0.05639147 ## 7 1 20 70.88067 0.7740865 1 NA NA 331 0.1543576 ## 8 1 20 70.88067 0.7740865 1 NA NA 348 0.3669095 ## 9 1 20 70.88067 0.7740865 1 NA NA 356 0.04364838 ## 10 1 20 70.88067 0.7740865 1 NA NA 368 0.01774773 ## # … with 54,890 more rowsLook at our variables
get_variables(lc)## [1] "b_Intercept" "b_age" "b_tumorsize" "b_lungcapacity" "b_age:tumorsize" "sd_did__Intercept" ## [7] "Intercept" "r_did[1,Intercept]" "r_did[2,Intercept]" "r_did[3,Intercept]" "r_did[4,Intercept]" "r_did[5,Intercept]" ## [13] "r_did[6,Intercept]" "r_did[7,Intercept]" "r_did[8,Intercept]" "r_did[9,Intercept]" "r_did[10,Intercept]" "r_did[11,Intercept]" ## [19] "r_did[12,Intercept]" "r_did[13,Intercept]" "r_did[14,Intercept]" "r_did[15,Intercept]" "r_did[16,Intercept]" "r_did[17,Intercept]" ## [25] "r_did[18,Intercept]" "r_did[19,Intercept]" "r_did[20,Intercept]" "r_did[21,Intercept]" "r_did[22,Intercept]" "r_did[23,Intercept]" ## [31] "r_did[24,Intercept]" "r_did[25,Intercept]" "r_did[26,Intercept]" "r_did[27,Intercept]" "r_did[28,Intercept]" "r_did[29,Intercept]" ## [37] "r_did[30,Intercept]" "r_did[31,Intercept]" "r_did[32,Intercept]" "r_did[33,Intercept]" "r_did[34,Intercept]" "r_did[35,Intercept]" ## [43] "r_did[36,Intercept]" "r_did[37,Intercept]" "r_did[38,Intercept]" "r_did[39,Intercept]" "r_did[40,Intercept]" "r_did[41,Intercept]" ## [49] "r_did[42,Intercept]" "r_did[43,Intercept]" "r_did[44,Intercept]" "r_did[45,Intercept]" "r_did[46,Intercept]" "r_did[47,Intercept]" ## [55] "r_did[48,Intercept]" "r_did[49,Intercept]" "r_did[50,Intercept]" "r_did[51,Intercept]" "r_did[52,Intercept]" "r_did[53,Intercept]" ## [61] "r_did[54,Intercept]" "r_did[55,Intercept]" "r_did[56,Intercept]" "r_did[57,Intercept]" "r_did[58,Intercept]" "r_did[59,Intercept]" ## [67] "r_did[60,Intercept]" "r_did[61,Intercept]" "r_did[62,Intercept]" "r_did[63,Intercept]" "r_did[64,Intercept]" "r_did[65,Intercept]" ## [73] "r_did[66,Intercept]" "r_did[67,Intercept]" "r_did[68,Intercept]" "r_did[69,Intercept]" "r_did[70,Intercept]" "r_did[71,Intercept]" ## [79] "r_did[72,Intercept]" "r_did[73,Intercept]" "r_did[74,Intercept]" "r_did[75,Intercept]" "r_did[76,Intercept]" "r_did[77,Intercept]" ## [85] "r_did[78,Intercept]" "r_did[79,Intercept]" "r_did[80,Intercept]" "r_did[81,Intercept]" "r_did[82,Intercept]" "r_did[83,Intercept]" ## [91] "r_did[84,Intercept]" "r_did[85,Intercept]" "r_did[86,Intercept]" "r_did[87,Intercept]" "r_did[88,Intercept]" "r_did[89,Intercept]" ## [97] "r_did[90,Intercept]" "r_did[91,Intercept]" "r_did[92,Intercept]" "r_did[93,Intercept]" "r_did[94,Intercept]" "r_did[95,Intercept]" ## [103] "r_did[96,Intercept]" "r_did[97,Intercept]" "r_did[98,Intercept]" "r_did[99,Intercept]" "r_did[100,Intercept]" "r_did[101,Intercept]"## [109] "r_did[102,Intercept]" "r_did[103,Intercept]" "r_did[104,Intercept]" "r_did[105,Intercept]" "r_did[106,Intercept]" "r_did[107,Intercept]"## [115] "r_did[108,Intercept]" "r_did[109,Intercept]" "r_did[110,Intercept]" "r_did[111,Intercept]" "r_did[112,Intercept]" "r_did[113,Intercept]"## [121] "r_did[114,Intercept]" "r_did[115,Intercept]" "r_did[116,Intercept]" "r_did[117,Intercept]" "r_did[118,Intercept]" "r_did[119,Intercept]"## [127] "r_did[120,Intercept]" "r_did[121,Intercept]" "r_did[122,Intercept]" "r_did[123,Intercept]" "r_did[124,Intercept]" "r_did[125,Intercept]"## [133] "r_did[126,Intercept]" "r_did[127,Intercept]" "r_did[128,Intercept]" "r_did[129,Intercept]" "r_did[130,Intercept]" "r_did[131,Intercept]"## [139] "r_did[132,Intercept]" "r_did[133,Intercept]" "r_did[134,Intercept]" "r_did[135,Intercept]" "r_did[136,Intercept]" "r_did[137,Intercept]"## [145] "r_did[138,Intercept]" "r_did[139,Intercept]" "r_did[140,Intercept]" "r_did[141,Intercept]" "r_did[142,Intercept]" "r_did[143,Intercept]"## [151] "r_did[144,Intercept]" "r_did[145,Intercept]" "r_did[146,Intercept]" "r_did[147,Intercept]" "r_did[148,Intercept]" "r_did[149,Intercept]"## [157] "r_did[150,Intercept]" "r_did[151,Intercept]" "r_did[152,Intercept]" "r_did[153,Intercept]" "r_did[154,Intercept]" "r_did[155,Intercept]"## [163] "r_did[156,Intercept]" "r_did[157,Intercept]" "r_did[158,Intercept]" "r_did[159,Intercept]" "r_did[160,Intercept]" "r_did[161,Intercept]"## [169] "r_did[162,Intercept]" "r_did[163,Intercept]" "r_did[164,Intercept]" "r_did[165,Intercept]" "r_did[166,Intercept]" "r_did[167,Intercept]"## [175] "r_did[168,Intercept]" "r_did[169,Intercept]" "r_did[170,Intercept]" "r_did[171,Intercept]" "r_did[172,Intercept]" "r_did[173,Intercept]"## [181] "r_did[174,Intercept]" "r_did[175,Intercept]" "r_did[176,Intercept]" "r_did[177,Intercept]" "r_did[178,Intercept]" "r_did[179,Intercept]"## [187] "r_did[180,Intercept]" "r_did[181,Intercept]" "r_did[182,Intercept]" "r_did[183,Intercept]" "r_did[184,Intercept]" "r_did[185,Intercept]"## [193] "r_did[186,Intercept]" "r_did[187,Intercept]" "r_did[188,Intercept]" "r_did[189,Intercept]" "r_did[190,Intercept]" "r_did[191,Intercept]"## [199] "r_did[192,Intercept]" "r_did[193,Intercept]" "r_did[194,Intercept]" "r_did[195,Intercept]" "r_did[196,Intercept]" "r_did[197,Intercept]"## [205] "r_did[198,Intercept]" "r_did[199,Intercept]" "r_did[200,Intercept]" "r_did[201,Intercept]" "r_did[202,Intercept]" "r_did[203,Intercept]"## [211] "r_did[204,Intercept]" "r_did[205,Intercept]" "r_did[206,Intercept]" "r_did[207,Intercept]" "r_did[208,Intercept]" "r_did[209,Intercept]"## [217] "r_did[210,Intercept]" "r_did[211,Intercept]" "r_did[212,Intercept]" "r_did[213,Intercept]" "r_did[214,Intercept]" "r_did[215,Intercept]"## [223] "r_did[216,Intercept]" "r_did[217,Intercept]" "r_did[218,Intercept]" "r_did[219,Intercept]" "r_did[220,Intercept]" "r_did[221,Intercept]"## [229] "r_did[222,Intercept]" "r_did[223,Intercept]" "r_did[224,Intercept]" "r_did[225,Intercept]" "r_did[226,Intercept]" "r_did[227,Intercept]"## [235] "r_did[228,Intercept]" "r_did[229,Intercept]" "r_did[230,Intercept]" "r_did[231,Intercept]" "r_did[232,Intercept]" "r_did[233,Intercept]"## [241] "r_did[234,Intercept]" "r_did[235,Intercept]" "r_did[236,Intercept]" "r_did[237,Intercept]" "r_did[238,Intercept]" "r_did[239,Intercept]"## [247] "r_did[240,Intercept]" "r_did[241,Intercept]" "r_did[242,Intercept]" "r_did[243,Intercept]" "r_did[244,Intercept]" "r_did[245,Intercept]"## [253] "r_did[246,Intercept]" "r_did[247,Intercept]" "r_did[248,Intercept]" "r_did[249,Intercept]" "r_did[250,Intercept]" "r_did[251,Intercept]"## [259] "r_did[252,Intercept]" "r_did[253,Intercept]" "r_did[254,Intercept]" "r_did[255,Intercept]" "r_did[256,Intercept]" "r_did[257,Intercept]"## [265] "r_did[258,Intercept]" "r_did[259,Intercept]" "r_did[260,Intercept]" "r_did[261,Intercept]" "r_did[262,Intercept]" "r_did[263,Intercept]"## [271] "r_did[264,Intercept]" "r_did[265,Intercept]" "r_did[266,Intercept]" "r_did[267,Intercept]" "r_did[268,Intercept]" "r_did[269,Intercept]"## [277] "r_did[270,Intercept]" "r_did[271,Intercept]" "r_did[272,Intercept]" "r_did[273,Intercept]" "r_did[274,Intercept]" "r_did[275,Intercept]"## [283] "r_did[276,Intercept]" "r_did[277,Intercept]" "r_did[278,Intercept]" "r_did[279,Intercept]" "r_did[280,Intercept]" "r_did[281,Intercept]"## [289] "r_did[282,Intercept]" "r_did[283,Intercept]" "r_did[284,Intercept]" "r_did[285,Intercept]" "r_did[286,Intercept]" "r_did[287,Intercept]"## [295] "r_did[288,Intercept]" "r_did[289,Intercept]" "r_did[290,Intercept]" "r_did[291,Intercept]" "r_did[292,Intercept]" "r_did[293,Intercept]"## [301] "r_did[294,Intercept]" "r_did[295,Intercept]" "r_did[296,Intercept]" "r_did[297,Intercept]" "r_did[298,Intercept]" "r_did[299,Intercept]"## [307] "r_did[300,Intercept]" "r_did[301,Intercept]" "r_did[302,Intercept]" "r_did[303,Intercept]" "r_did[304,Intercept]" "r_did[305,Intercept]"## [313] "r_did[306,Intercept]" "r_did[307,Intercept]" "r_did[308,Intercept]" "r_did[309,Intercept]" "r_did[310,Intercept]" "r_did[311,Intercept]"## [319] "r_did[312,Intercept]" "r_did[313,Intercept]" "r_did[314,Intercept]" "r_did[315,Intercept]" "r_did[316,Intercept]" "r_did[317,Intercept]"## [325] "r_did[318,Intercept]" "r_did[319,Intercept]" "r_did[320,Intercept]" "r_did[321,Intercept]" "r_did[322,Intercept]" "r_did[323,Intercept]"## [331] "r_did[324,Intercept]" "r_did[325,Intercept]" "r_did[326,Intercept]" "r_did[327,Intercept]" "r_did[328,Intercept]" "r_did[329,Intercept]"## [337] "r_did[330,Intercept]" "r_did[331,Intercept]" "r_did[332,Intercept]" "r_did[333,Intercept]" "r_did[334,Intercept]" "r_did[335,Intercept]"## [343] "r_did[336,Intercept]" "r_did[337,Intercept]" "r_did[338,Intercept]" "r_did[339,Intercept]" "r_did[340,Intercept]" "r_did[341,Intercept]"## [349] "r_did[342,Intercept]" "r_did[343,Intercept]" "r_did[344,Intercept]" "r_did[345,Intercept]" "r_did[346,Intercept]" "r_did[347,Intercept]"## [355] "r_did[348,Intercept]" "r_did[349,Intercept]" "r_did[350,Intercept]" "r_did[351,Intercept]" "r_did[352,Intercept]" "r_did[353,Intercept]"## [361] "r_did[354,Intercept]" "r_did[355,Intercept]" "r_did[356,Intercept]" "r_did[357,Intercept]" "r_did[358,Intercept]" "r_did[359,Intercept]"## [367] "r_did[360,Intercept]" "r_did[361,Intercept]" "r_did[362,Intercept]" "r_did[363,Intercept]" "r_did[364,Intercept]" "r_did[365,Intercept]"## [373] "r_did[366,Intercept]" "r_did[367,Intercept]" "r_did[368,Intercept]" "r_did[369,Intercept]" "r_did[370,Intercept]" "r_did[371,Intercept]"## [379] "r_did[372,Intercept]" "r_did[373,Intercept]" "r_did[374,Intercept]" "r_did[375,Intercept]" "r_did[376,Intercept]" "r_did[377,Intercept]"## [385] "r_did[378,Intercept]" "r_did[379,Intercept]" "r_did[380,Intercept]" "r_did[381,Intercept]" "r_did[382,Intercept]" "r_did[383,Intercept]"## [391] "r_did[384,Intercept]" "r_did[385,Intercept]" "r_did[386,Intercept]" "r_did[387,Intercept]" "r_did[388,Intercept]" "r_did[389,Intercept]"## [397] "r_did[390,Intercept]" "r_did[391,Intercept]" "r_did[392,Intercept]" "r_did[393,Intercept]" "r_did[394,Intercept]" "r_did[395,Intercept]"## [403] "r_did[396,Intercept]" "r_did[397,Intercept]" "r_did[398,Intercept]" "r_did[399,Intercept]" "r_did[400,Intercept]" "r_did[401,Intercept]"## [409] "r_did[402,Intercept]" "r_did[403,Intercept]" "r_did[404,Intercept]" "r_did[405,Intercept]" "r_did[406,Intercept]" "r_did[407,Intercept]"## [415] "lp__" "z_1[1,1]" "z_1[1,2]" "z_1[1,3]" "z_1[1,4]" "z_1[1,5]" ## [421] "z_1[1,6]" "z_1[1,7]" "z_1[1,8]" "z_1[1,9]" "z_1[1,10]" "z_1[1,11]" ## [427] "z_1[1,12]" "z_1[1,13]" "z_1[1,14]" "z_1[1,15]" "z_1[1,16]" "z_1[1,17]" ## [433] "z_1[1,18]" "z_1[1,19]" "z_1[1,20]" "z_1[1,21]" "z_1[1,22]" "z_1[1,23]" ## [439] "z_1[1,24]" "z_1[1,25]" "z_1[1,26]" "z_1[1,27]" "z_1[1,28]" "z_1[1,29]" ## [445] "z_1[1,30]" "z_1[1,31]" "z_1[1,32]" "z_1[1,33]" "z_1[1,34]" "z_1[1,35]" ## [451] "z_1[1,36]" "z_1[1,37]" "z_1[1,38]" "z_1[1,39]" "z_1[1,40]" "z_1[1,41]" ## [457] "z_1[1,42]" "z_1[1,43]" "z_1[1,44]" "z_1[1,45]" "z_1[1,46]" "z_1[1,47]" ## [463] "z_1[1,48]" "z_1[1,49]" "z_1[1,50]" "z_1[1,51]" "z_1[1,52]" "z_1[1,53]" ## [469] "z_1[1,54]" "z_1[1,55]" "z_1[1,56]" "z_1[1,57]" "z_1[1,58]" "z_1[1,59]" ## [475] "z_1[1,60]" "z_1[1,61]" "z_1[1,62]" "z_1[1,63]" "z_1[1,64]" "z_1[1,65]" ## [481] "z_1[1,66]" "z_1[1,67]" "z_1[1,68]" "z_1[1,69]" "z_1[1,70]" "z_1[1,71]" ## [487] "z_1[1,72]" "z_1[1,73]" "z_1[1,74]" "z_1[1,75]" "z_1[1,76]" "z_1[1,77]" ## [493] "z_1[1,78]" "z_1[1,79]" "z_1[1,80]" "z_1[1,81]" "z_1[1,82]" "z_1[1,83]" ## [499] "z_1[1,84]" "z_1[1,85]" "z_1[1,86]" "z_1[1,87]" "z_1[1,88]" "z_1[1,89]" ## [505] "z_1[1,90]" "z_1[1,91]" "z_1[1,92]" "z_1[1,93]" "z_1[1,94]" "z_1[1,95]" ## [511] "z_1[1,96]" "z_1[1,97]" "z_1[1,98]" "z_1[1,99]" "z_1[1,100]" "z_1[1,101]" ## [517] "z_1[1,102]" "z_1[1,103]" "z_1[1,104]" "z_1[1,105]" "z_1[1,106]" "z_1[1,107]" ## [523] "z_1[1,108]" "z_1[1,109]" "z_1[1,110]" "z_1[1,111]" "z_1[1,112]" "z_1[1,113]" ## [529] "z_1[1,114]" "z_1[1,115]" "z_1[1,116]" "z_1[1,117]" "z_1[1,118]" "z_1[1,119]" ## [535] "z_1[1,120]" "z_1[1,121]" "z_1[1,122]" "z_1[1,123]" "z_1[1,124]" "z_1[1,125]" ## [541] "z_1[1,126]" "z_1[1,127]" "z_1[1,128]" "z_1[1,129]" "z_1[1,130]" "z_1[1,131]" ## [547] "z_1[1,132]" "z_1[1,133]" "z_1[1,134]" "z_1[1,135]" "z_1[1,136]" "z_1[1,137]" ## [553] "z_1[1,138]" "z_1[1,139]" "z_1[1,140]" "z_1[1,141]" "z_1[1,142]" "z_1[1,143]" ## [559] "z_1[1,144]" "z_1[1,145]" "z_1[1,146]" "z_1[1,147]" "z_1[1,148]" "z_1[1,149]" ## [565] "z_1[1,150]" "z_1[1,151]" "z_1[1,152]" "z_1[1,153]" "z_1[1,154]" "z_1[1,155]" ## [571] "z_1[1,156]" "z_1[1,157]" "z_1[1,158]" "z_1[1,159]" "z_1[1,160]" "z_1[1,161]" ## [577] "z_1[1,162]" "z_1[1,163]" "z_1[1,164]" "z_1[1,165]" "z_1[1,166]" "z_1[1,167]" ## [583] "z_1[1,168]" "z_1[1,169]" "z_1[1,170]" "z_1[1,171]" "z_1[1,172]" "z_1[1,173]" ## [589] "z_1[1,174]" "z_1[1,175]" "z_1[1,176]" "z_1[1,177]" "z_1[1,178]" "z_1[1,179]" ## [595] "z_1[1,180]" "z_1[1,181]" "z_1[1,182]" "z_1[1,183]" "z_1[1,184]" "z_1[1,185]" ## [601] "z_1[1,186]" "z_1[1,187]" "z_1[1,188]" "z_1[1,189]" "z_1[1,190]" "z_1[1,191]" ## [607] "z_1[1,192]" "z_1[1,193]" "z_1[1,194]" "z_1[1,195]" "z_1[1,196]" "z_1[1,197]" ## [613] "z_1[1,198]" "z_1[1,199]" "z_1[1,200]" "z_1[1,201]" "z_1[1,202]" "z_1[1,203]" ## [619] "z_1[1,204]" "z_1[1,205]" "z_1[1,206]" "z_1[1,207]" "z_1[1,208]" "z_1[1,209]" ## [625] "z_1[1,210]" "z_1[1,211]" "z_1[1,212]" "z_1[1,213]" "z_1[1,214]" "z_1[1,215]" ## [631] "z_1[1,216]" "z_1[1,217]" "z_1[1,218]" "z_1[1,219]" "z_1[1,220]" "z_1[1,221]" ## [637] "z_1[1,222]" "z_1[1,223]" "z_1[1,224]" "z_1[1,225]" "z_1[1,226]" "z_1[1,227]" ## [643] "z_1[1,228]" "z_1[1,229]" "z_1[1,230]" "z_1[1,231]" "z_1[1,232]" "z_1[1,233]" ## [649] "z_1[1,234]" "z_1[1,235]" "z_1[1,236]" "z_1[1,237]" "z_1[1,238]" "z_1[1,239]" ## [655] "z_1[1,240]" "z_1[1,241]" "z_1[1,242]" "z_1[1,243]" "z_1[1,244]" "z_1[1,245]" ## [661] "z_1[1,246]" "z_1[1,247]" "z_1[1,248]" "z_1[1,249]" "z_1[1,250]" "z_1[1,251]" ## [667] "z_1[1,252]" "z_1[1,253]" "z_1[1,254]" "z_1[1,255]" "z_1[1,256]" "z_1[1,257]" ## [673] "z_1[1,258]" "z_1[1,259]" "z_1[1,260]" "z_1[1,261]" "z_1[1,262]" "z_1[1,263]" ## [679] "z_1[1,264]" "z_1[1,265]" "z_1[1,266]" "z_1[1,267]" "z_1[1,268]" "z_1[1,269]" ## [685] "z_1[1,270]" "z_1[1,271]" "z_1[1,272]" "z_1[1,273]" "z_1[1,274]" "z_1[1,275]" ## [691] "z_1[1,276]" "z_1[1,277]" "z_1[1,278]" "z_1[1,279]" "z_1[1,280]" "z_1[1,281]" ## [697] "z_1[1,282]" "z_1[1,283]" "z_1[1,284]" "z_1[1,285]" "z_1[1,286]" "z_1[1,287]" ## [703] "z_1[1,288]" "z_1[1,289]" "z_1[1,290]" "z_1[1,291]" "z_1[1,292]" "z_1[1,293]" ## [709] "z_1[1,294]" "z_1[1,295]" "z_1[1,296]" "z_1[1,297]" "z_1[1,298]" "z_1[1,299]" ## [715] "z_1[1,300]" "z_1[1,301]" "z_1[1,302]" "z_1[1,303]" "z_1[1,304]" "z_1[1,305]" ## [721] "z_1[1,306]" "z_1[1,307]" "z_1[1,308]" "z_1[1,309]" "z_1[1,310]" "z_1[1,311]" ## [727] "z_1[1,312]" "z_1[1,313]" "z_1[1,314]" "z_1[1,315]" "z_1[1,316]" "z_1[1,317]" ## [733] "z_1[1,318]" "z_1[1,319]" "z_1[1,320]" "z_1[1,321]" "z_1[1,322]" "z_1[1,323]" ## [739] "z_1[1,324]" "z_1[1,325]" "z_1[1,326]" "z_1[1,327]" "z_1[1,328]" "z_1[1,329]" ## [745] "z_1[1,330]" "z_1[1,331]" "z_1[1,332]" "z_1[1,333]" "z_1[1,334]" "z_1[1,335]" ## [751] "z_1[1,336]" "z_1[1,337]" "z_1[1,338]" "z_1[1,339]" "z_1[1,340]" "z_1[1,341]" ## [757] "z_1[1,342]" "z_1[1,343]" "z_1[1,344]" "z_1[1,345]" "z_1[1,346]" "z_1[1,347]" ## [763] "z_1[1,348]" "z_1[1,349]" "z_1[1,350]" "z_1[1,351]" "z_1[1,352]" "z_1[1,353]" ## [769] "z_1[1,354]" "z_1[1,355]" "z_1[1,356]" "z_1[1,357]" "z_1[1,358]" "z_1[1,359]" ## [775] "z_1[1,360]" "z_1[1,361]" "z_1[1,362]" "z_1[1,363]" "z_1[1,364]" "z_1[1,365]" ## [781] "z_1[1,366]" "z_1[1,367]" "z_1[1,368]" "z_1[1,369]" "z_1[1,370]" "z_1[1,371]" ## [787] "z_1[1,372]" "z_1[1,373]" "z_1[1,374]" "z_1[1,375]" "z_1[1,376]" "z_1[1,377]" ## [793] "z_1[1,378]" "z_1[1,379]" "z_1[1,380]" "z_1[1,381]" "z_1[1,382]" "z_1[1,383]" ## [799] "z_1[1,384]" "z_1[1,385]" "z_1[1,386]" "z_1[1,387]" "z_1[1,388]" "z_1[1,389]" ## [805] "z_1[1,390]" "z_1[1,391]" "z_1[1,392]" "z_1[1,393]" "z_1[1,394]" "z_1[1,395]" ## [811] "z_1[1,396]" "z_1[1,397]" "z_1[1,398]" "z_1[1,399]" "z_1[1,400]" "z_1[1,401]" ## [817] "z_1[1,402]" "z_1[1,403]" "z_1[1,404]" "z_1[1,405]" "z_1[1,406]" "z_1[1,407]" ## [823] "accept_stat__" "treedepth__" "stepsize__" "divergent__" "n_leapfrog__" "energy__"Get all draws: Intercept
Notice I'm using spread_draws() here for a slightly different output format
int <- lc %>% spread_draws(b_Intercept)int## # A tibble: 4,000 x 4## .chain .iteration .draw b_Intercept## <int> <int> <int> <dbl>## 1 1 1 1 0.799584 ## 2 1 2 2 -0.0137843## 3 1 3 3 1.11889 ## 4 1 4 4 2.99813 ## 5 1 5 5 1.37627 ## 6 1 6 6 2.57053 ## 7 1 7 7 3.96603 ## 8 1 8 8 3.98211 ## 9 1 9 9 0.561609 ## 10 1 10 10 0.135172 ## # … with 3,990 more rowsGrab random effects
- The random effect name is
r_did
spread_draws(lc, r_did[did, term])## # A tibble: 1,628,000 x 6## # Groups: did, term [407]## did term r_did .chain .iteration .draw## <int> <chr> <dbl> <int> <int> <int>## 1 1 Intercept -1.94541 1 1 1## 2 1 Intercept -8.03429 1 2 2## 3 1 Intercept -3.57734 1 3 3## 4 1 Intercept -2.27834 1 4 4## 5 1 Intercept -3.41082 1 5 5## 6 1 Intercept -2.17562 1 6 6## 7 1 Intercept -3.63758 1 7 7## 8 1 Intercept -2.9472 1 8 8## 9 1 Intercept -2.66865 1 9 9## 10 1 Intercept -5.06383 1 10 10## # … with 1,627,990 more rowsLook at did distributions
First 75 doctors
dids <- spread_draws(lc, r_did[did, ]) # all terms, which is just onedids %>% ungroup() %>% filter(did %in% 1:75) %>% mutate(did = fct_reorder(factor(did), r_did)) %>% ggplot(aes(x = r_did, y = factor(did))) + ggridges::geom_density_ridges(color = NA, fill = "#61adff") + theme(panel.grid.major.y = element_blank(), panel.grid.minor = element_blank(), axis.text.y = element_blank())Long format
Use gather_draws() for a longer format (as we did before)
fixed_l <- lc %>% gather_draws(b_Intercept, b_age, b_tumorsize, b_lungcapacity, `b_age:tumorsize`)fixed_l## # A tibble: 20,000 x 5## # Groups: .variable [5]## .chain .iteration .draw .variable .value## <int> <int> <int> <chr> <dbl>## 1 1 1 1 b_Intercept 0.799584 ## 2 1 2 2 b_Intercept -0.0137843## 3 1 3 3 b_Intercept 1.11889 ## 4 1 4 4 b_Intercept 2.99813 ## 5 1 5 5 b_Intercept 1.37627 ## 6 1 6 6 b_Intercept 2.57053 ## 7 1 7 7 b_Intercept 3.96603 ## 8 1 8 8 b_Intercept 3.98211 ## 9 1 9 9 b_Intercept 0.561609 ## 10 1 10 10 b_Intercept 0.135172 ## # … with 19,990 more rowsMultiple comparisons
One of the nicest things about Bayes is that any comparison you want to make can be made without jumping through a lot of additional hoops (e.g., adjusting \(\alpha\)).
Scenario
Imagine a 35 year old has a tumor measuring 58 millimeters and a lung capacity rating of 0.81.
Multiple comparisons
One of the nicest things about Bayes is that any comparison you want to make can be made without jumping through a lot of additional hoops (e.g., adjusting \(\alpha\)).
Scenario
Imagine a 35 year old has a tumor measuring 58 millimeters and a lung capacity rating of 0.81.
What would we estimate as the probability of remission if this patient had did == 1 versus did == 2?
Fixed effects
Not really "fixed", but rather just average relation
fe <- lc %>% spread_draws(b_Intercept, b_age, b_tumorsize, b_lungcapacity, `b_age:tumorsize`)fe## # A tibble: 4,000 x 8## .chain .iteration .draw b_Intercept b_age b_tumorsize b_lungcapacity `b_age:tumorsize`## <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 1 1 0.799584 -0.0318583 0.00418452 0.179215 -0.000279332## 2 1 2 2 -0.0137843 -0.016284 0.0151118 0.043369 -0.000475806## 3 1 3 3 1.11889 -0.0304524 0.00133416 0.047453 -0.000306942## 4 1 4 4 2.99813 -0.0769063 -0.0199561 0.157362 0.000205206## 5 1 5 5 1.37627 -0.0447789 0.00165146 0.204612 -0.000215956## 6 1 6 6 2.57053 -0.0661329 -0.0211792 0.161738 0.000200291## 7 1 7 7 3.96603 -0.0934501 -0.0318102 0.12162 0.000429057## 8 1 8 8 3.98211 -0.090909 -0.0310474 -0.0166011 0.000409157## 9 1 9 9 0.561609 -0.0246561 0.00422727 0.161216 -0.000302065## 10 1 10 10 0.135172 -0.0192131 0.0167028 -0.00243727 -0.00050405 ## # … with 3,990 more rowsData
age <- 35tumor_size <- 58lung_cap <- 0.81Population-level predictions (there's other ways we could do this, but it's good to remind ourselves the "by hand" version too)
pop_level <- fe$b_Intercept + (fe$b_age * age) + (fe$b_tumorsize * tumor_size) + (fe$b_lungcapacity * lung_cap) + (fe$`b_age:tumorsize` * (age * tumor_size))pop_level## [1] -0.49463415 -0.63799719 -0.45421805 -0.30701290 -0.36786178 -0.43491659 -0.18021719 -0.18331238 -0.53877983 -0.59371979 -0.42843066 -0.50226066## [13] -0.51347752 -0.35731807 -0.30377370 -0.45074384 -0.39412555 -0.42227993 -0.55596505 -0.26324322 -0.37179408 -0.28712718 -0.43175350 -0.49671339## [25] -0.41777911 -0.24052536 -0.26502943 -0.14023521 -0.08610160 -0.25673816 -0.15608123 -0.35687678 -0.48132277 -0.25568533 -0.30806044 -0.26857323## [37] -0.39127941 -0.54278890 -0.81346645 -0.82226173 -0.69583974 -0.50434511 -0.51757279 -0.42181277 -0.46060622 -0.47654284 -0.67076151 -0.52865674## [49] -0.50703059 -0.55960140 -0.72656231 -0.85642544 -0.59342675 -0.50563625 -0.69911641 -0.62921079 -0.47659289 -0.36797823 -0.52780246 -0.56145785## [61] -0.47342652 -0.43957181 -0.25880691 -0.59541750 -0.39725010 -0.43700704 -0.32979246 -0.48930002 -0.34037838 -0.42941843 -0.37282981 -0.46057624## [73] -0.37872082 -0.30499434 -0.18936291 -0.16394375 -0.16392711 -0.33959043 -0.43532333 -0.38244622 -0.55614286 -0.42982285 -0.58842529 -0.41600740## [85] -0.30694046 -0.34863954 -0.36616642 -0.40719484 -0.19466787 -0.52755250 -0.39260994 -0.42396988 -0.33853851 -0.63979403 -0.65107885 -0.63234653## [97] -0.44115504 -0.50257323 -0.48584442 -0.39829851 -0.52682195 -0.75393167 -0.64291019 -0.40275983 -0.46012324 -0.40830093 -0.27680348 -0.36291805## [109] -0.38486662 -0.30792298 -0.49470714 -0.35194097 -0.53235345 -0.39987262 -0.62171693 -0.59885076 -0.15310728 -0.52821731 -0.55486560 -0.56943779## [121] -0.49320855 -0.60748829 -0.75971199 -0.63040555 -0.29346239 -0.48582743 -0.35358205 -0.58025209 -0.63223742 -0.65972712 -0.41111676 -0.46430197## [133] -0.40270796 -0.20372941 -0.26266338 -0.36569821 -0.65898268 -0.44212342 -0.47130629 -0.38250941 -0.37678517 -0.27351627 -0.31134589 -0.45595068## [145] -0.44170347 -0.28810830 -0.48145920 -0.27903673 -0.78540349 -0.37470447 -0.47767079 -0.37265653 -0.49826104 -0.34496852 -0.54122600 -0.56408039## [157] -0.50511392 -0.54310512 -0.66747176 -0.56169970 -0.51106171 -0.60235596 -0.41492964 -0.62965281 -0.43045092 -0.40740492 -0.62226800 -0.83843865## [169] -0.43630412 -0.62989361 -0.55276582 -0.54362410 -0.48518853 -0.70920281 -0.85427974 -0.96549777 -0.73809323 -0.49057462 -0.77237772 -0.69287479## [181] -0.80746374 -0.69359188 -0.67467258 -0.29285036 -0.45623888 -0.73690692 -0.40663800 -0.40522645 -0.42455963 -0.21786731 -0.22797657 -0.16852969## [193] -0.61977584 -0.59170300 -0.65788470 -0.42483561 -0.30915285 -0.36082242 -0.30427860 -0.27281219 -0.38456528 -0.53237083 -0.55168160 -0.47085094## [205] -0.15359036 -0.51205204 -0.67046759 -0.37497836 -0.42920173 -0.35699251 -0.55938641 -0.55247006 -0.20282739 -0.09109705 -0.17101543 -0.30788548## [217] -0.26681868 -0.28089481 -0.35880673 -0.38819741 -0.37491515 -0.37880972 -0.36462283 -0.38583089 -0.39988731 -0.56301305 -0.29158540 -0.69860686## [229] -0.63010749 -0.65778583 -0.61695614 -0.62030029 -0.53221555 -0.80351954 -0.74231636 -0.58150913 -0.66949356 -0.38617209 -0.55025464 -0.55920178## [241] -0.57606234 -0.32912555 -0.66729190 -0.59760456 -0.52889752 -0.54748383 -0.76495828 -0.44575049 -0.39556809 -0.32352230 -0.53083774 -0.51985977## [253] -0.63351699 -0.52139350 -0.68703855 -0.61788860 -0.60749694 -0.68485857 -0.60938959 -0.35428111 -0.38430916 -0.47471253 -0.47156081 -0.64829922## [265] -0.38025083 -0.34003693 -0.44803354 -0.40024876 -0.34706353 -0.39502133 -0.34563470 -0.49499641 -0.46353489 -0.50240763 -0.65381740 -0.59325556## [277] -0.47191805 -0.31803397 -0.41373397 -0.64270044 -0.38888001 -0.63043172 -0.43200623 -0.55135738 -0.64940268 -0.69600095 -0.67096432 -0.69830310## [289] -0.63651296 -0.50519284 -0.29161781 -0.34888131 -0.21304204 -0.38896816 -0.55735573 -0.58117885 -0.23441916 -0.49017234 -0.38023746 -0.54269242## [301] -0.04702058 -0.31218006 -0.34781871 -0.31661353 -0.20891339 -0.51448583 -0.43224889 -0.51795320 -0.30931029 -0.28951744 -0.32678569 -0.57193843## [313] -0.47267478 -0.35203033 -0.22963899 -0.55353163 -0.39745223 -0.48944921 -0.57216353 -0.40719847 -0.68853569 -0.63286432 -0.57334890 -0.57059552## [325] -0.44440313 -0.54202901 -0.50901253 -0.24621145 -0.50073500 -0.27580690 -0.45731987 -0.25640227 -0.24825424 -0.44030979 -0.28847905 -0.20820445## [337] -0.25970028 -0.34224445 -0.40493506 -0.39409035 -0.36685220 -0.27554787 -0.17217499 -0.07451407 -0.50998388 -0.54910472 -0.56380956 -0.43169093## [349] -0.62289513 -0.45906294 -0.26239654 -0.13949602 -0.22103531 -0.19418731 -0.43158266 -0.30010798 -0.60828603 -0.37305323 -0.22257543 -0.51375589## [361] -0.53980475 -0.53871610 -0.59130677 -0.36926341 -0.48915296 -0.43416343 -0.47392145 -0.61364284 -0.63093786 -0.53943439 -0.60144042 -0.57304569## [373] -0.49351463 -0.68450893 -0.58299145 -0.66301785 -0.63279105 -0.68165315 -0.47106015 -0.80247013 -0.44477521 -0.57534662 -0.59617539 -0.76202292## [385] -0.33172144 -0.58116086 -0.41514650 -0.27376817 -0.35436144 -0.60307242 -0.44091927 -0.30169729 -0.44807187 -0.58157569 -0.61934046 -0.69706444## [397] -0.61476282 -0.41326292 -0.52581169 -0.39711389 -0.33718128 -0.35206145 -0.35116984 -0.38783572 -0.41965731 -0.59395268 -0.37076536 -0.62092013## [409] -0.57777531 -0.69923430 -0.58605528 -0.35464895 -0.61352792 -0.60731615 -0.58652451 -0.46485088 -0.53232830 -0.45761893 -0.47945821 -0.26470046## [421] -0.29404571 -0.54922361 -0.47947931 -0.30976861 -0.56834877 -0.50786444 -0.64750516 -0.50158309 -0.62275239 -0.47413783 -0.42639606 -0.70912867## [433] -0.67644189 -0.52042603 -0.54865398 -0.56546109 -0.60075379 -0.48165397 -0.44021508 -0.41028918 -0.34425476 -0.44470954 -0.58348292 -0.46224060## [445] -0.30756695 -0.22806762 -0.33462586 -0.39240444 -0.35806456 -0.35151860 -0.44635260 -0.17930971 -0.09295412 -0.56570445 -0.11568823 -0.10215407## [457] -0.36221043 -0.53042851 -0.47709880 -0.27447590 -0.32864854 -0.50720563 -0.50156452 -0.24638333 -0.33070239 -0.36218839 -0.35262833 -0.53412647## [469] -0.42185428 -0.48144896 -0.37782697 -0.84994152 -0.66606604 -0.44723318 -0.51287825 -0.60897554 -0.25657120 -0.45148677 -0.52876387 -0.29873955## [481] -0.41941900 -0.44630641 -0.60580220 -0.30491773 -0.41726304 -0.44708365 -0.48888179 -0.70425650 -0.36852671 -0.46878764 -0.45414330 -0.33564519## [493] -0.38554253 -0.30876338 -0.46376092 -0.48143980 -0.47450597 -0.33052645 -0.39197300 -0.11322858 -0.40469383 -0.60585254 -0.78228472 -0.56576451## [505] -0.41221155 -0.12059208 -0.73820555 -0.70430338 -0.60320736 -0.61766330 -0.35360752 -0.40933106 -0.18994831 -0.57014846 -0.72442497 -0.28426097## [517] -0.51075093 -0.68753599 -0.80317598 -0.39721059 -0.67376878 -0.36880685 -0.51550835 -0.21227700 -0.57546195 -0.43166261 -0.61215164 -0.29931902## [529] -0.51060121 -0.30659173 -0.42946742 -0.54318498 -0.74528464 -0.46725178 -0.60824403 -0.72944867 -0.66553637 -0.87184163 -0.72119754 -0.48266016## [541] -0.65064765 -0.64149654 -0.46404278 -0.63054578 -0.34110771 -0.50215793 -0.47009774 -0.38356081 -0.21967669 -0.50668294 -0.41478749 -0.39650861## [553] -0.64931286 -0.66535992 -0.24352702 -0.34237658 -0.43879145 -0.44612567 -0.52531153 -0.71399685 -0.59657936 -0.55415477 -0.53627298 -0.55298671## [565] -0.26113149 -0.56915518 -0.11338429 -0.30301345 -0.36309790 -0.36939736 -0.47548134 -0.29037600 -0.04821158 -0.24513035 -0.47309227 -0.31317579## [577] -0.55067801 -0.33424785 -0.48751513 -0.56565354 -0.58155307 -0.62186861 -0.34379716 -0.34779879 -0.21029787 -0.48478574 -1.06659256 -0.68005719## [589] -0.57330614 -0.45636422 -0.48602390 -0.49015495 -0.46735291 -0.43512316 -0.47599465 -0.40433338 -0.33262144 -0.33923400 -0.14055196 -0.29461275## [601] -0.41703349 -0.47515022 -0.37411465 -0.53454904 -0.32378179 -0.28753756 -0.57111165 -0.44873340 -0.39247133 -0.36187798 -0.59162624 -0.43918013## [613] -0.63760317 -0.97033558 -0.84496979 -0.98974054 -0.90273369 -0.92243013 -0.54073896 -0.71979234 -0.49862966 -0.69917444 -0.61896356 -0.89836352## [625] -0.75234593 -0.50797009 -0.56539005 -0.48485376 -0.57902114 -0.77908339 -0.73572188 -0.37142164 -0.58034174 -0.55245280 -0.80392852 -0.54995018## [637] -0.32053428 -0.39599662 -0.35534151 -0.52953679 -0.25239436 -0.42110525 -0.39014112 -0.63080139 -0.48381685 -0.54279696 -0.64413521 -0.56785077## [649] -0.76679967 -0.41257218 -0.54188408 -0.16545839 -0.20502355 -0.53742959 -0.65372835 -0.41676803 -0.30209008 -0.36773323 -0.52769280 -0.46142819## [661] -0.38247678 -0.32347078 -0.81170113 -0.58588446 -0.80074933 -0.64639607 -0.77787106 -0.59002533 -0.44644290 -0.40446554 -0.44594037 -0.30802340## [673] -0.48799144 -0.30937898 -0.41280775 -0.39891460 -0.36213856 -0.26463790 -0.51542691 -0.35867985 -0.51571893 -0.33072878 -0.49233675 -0.43238566## [685] -0.52855070 -0.30284529 -0.39832969 -0.42581869 -0.51765707 -0.53495231 -0.48056346 -0.38714798 -0.47508177 -0.44815678 -0.30936910 -0.58520166## [697] -0.42547098 -0.39084422 -0.64252206 -0.66891927 -0.54915497 -0.40870166 -0.47891268 -0.57959249 -0.53781088 -0.32170069 -0.38345359 -0.48867374## [709] -0.28631876 -0.27439130 -0.28121342 -0.28547182 -0.31044153 -0.31849001 -0.20016367 -0.19175966 -0.67651170 -0.56139520 -0.44387533 -0.52851261## [721] -0.31578971 -0.36867332 -0.19064869 -0.43118721 -0.39501664 -0.31506283 -0.47706654 -0.48299738 -0.60153616 -0.49833489 -0.89726796 -0.64677084## [733] -0.68106163 -0.49002018 -0.42345753 -0.48967475 -0.49557856 -0.31202057 -0.46126563 -0.40645016 -0.47221346 -0.43786080 -0.26694004 -0.71580842## [745] -0.66390108 -0.66950119 -0.73497821 -0.28194838 -0.43082623 -0.62804370 -0.58684122 -0.35172264 -0.40328295 -0.20237317 -0.30327210 -0.02538725## [757] -0.29886391 -0.35424189 -0.75967165 -0.54613824 -0.52383545 -0.57394735 -0.67669953 -0.67540541 -0.28365207 -0.31915277 -0.41455799 -0.37444493## [769] -0.39431352 -0.46126736 -0.54998338 -0.56009666 -0.54010148 -0.55985192 -0.44234394 -0.35101783 -0.57107176 -0.51651857 -0.56053009 -0.56549531## [781] -0.32370711 -0.44608730 -0.47216121 -0.57018037 -0.50352304 -0.31605559 -0.48825816 -0.48613089 -0.40250550 -0.54074320 -0.38751757 -0.52800491## [793] -0.38042384 -0.24347600 -0.15731675 -0.24039539 -0.17533492 -0.12381527 -0.39718746 -0.54410888 -0.36852454 -0.40885739 -0.52772986 -0.66979442## [805] -0.53835701 -0.38394666 -0.52180128 -0.39350474 -0.70564016 -0.62740319 -0.55362892 -0.59560654 -0.53910962 -0.51376774 -0.52371985 -0.65458841## [817] -0.44586077 -0.43309747 -0.45220011 -0.18855500 -0.42611938 -0.29801128 -0.26318284 -0.38151842 -0.37169036 -0.46178289 -0.36486204 -0.42841791## [829] -0.47475949 -0.50045261 -0.41019831 -0.58062327 -0.62513651 -0.63199364 -0.35886294 -0.54489097 -0.56245103 -0.40228191 -0.47927573 -0.44615842## [841] -0.62676808 -0.35500107 -0.48838685 -0.37075933 -0.66025610 -0.74470274 -0.67668657 -0.74758663 -0.53852126 -0.73965334 -0.62903476 -0.48446313## [853] -0.70790595 -0.40182184 -0.36795590 -0.34225104 -0.51745789 -0.27518504 -0.43902968 -0.43441410 -0.55663059 -0.62526180 -0.56721559 -0.52690638## [865] -0.67068822 -0.60977886 -0.61100614 -0.43823814 -0.33655155 -0.31021006 -0.40288928 -0.51621584 -0.34298110 -0.33027040 -0.46727286 -0.39160283## [877] -0.41321720 -0.40470673 -0.14565986 -0.38852166 -0.49802266 -0.28141874 -0.43673993 -0.98229539 -0.69876277 -0.41448127 -0.51708848 -0.52341293## [889] -0.69194932 -0.20588577 -0.63641611 -0.53135215 -0.40028891 -0.19697771 -0.27935194 -0.14633638 -0.47186954 -0.42618852 -0.40402895 -0.44116150## [901] -0.55425942 -0.72548817 -0.58779297 -0.44790284 -0.31016077 -0.23906778 -0.41653599 -0.20572668 -0.33573534 -0.28056454 -0.26038494 -0.20989298## [913] -0.46705026 -0.43113601 -0.39685455 -0.47062546 -0.42734396 -0.29048728 -0.37472762 -0.13789888 -0.42075641 -0.33858749 -0.25639988 -0.38734437## [925] -0.03055166 -0.30637544 -0.45966453 -0.41933959 -0.13554882 -0.67428474 -0.53492480 -0.48632755 -0.33576085 -0.57144915 -0.62605650 -0.69489913## [937] -0.74932929 -0.60625094 -0.66873166 -0.88704353 -0.57023319 -0.60645862 -0.51997354 -0.57393974 -0.50000027 -0.55741983 -0.41119988 -0.48956074## [949] -0.75032016 -0.52647771 -0.56779293 -0.78810987 -0.66621734 -0.68661438 -0.59592653 -0.54561419 -0.31018503 -0.50593670 -0.58981273 -0.38307070## [961] -0.57748734 -0.48419221 -0.40374087 -0.33072821 -0.20559085 -0.20817144 -0.68007571 -0.41190176 -0.47717614 -0.49681309 -0.57200883 -0.50615485## [973] -0.52272186 -0.59463991 -0.48307741 -0.73904469 -0.45707404 -0.68563076 -0.23489347 -0.93192180 -0.64131414 -0.53887316 -0.67767751 -0.10956105## [985] -0.24128288 -0.51787362 -0.18199276 -0.54136341 -0.18852362 -0.38639066 -0.40628911 -0.14541143 -0.48937661 -0.60651916 -0.51236978 -0.51489665## [997] -0.38909958 -0.67320141 -0.69647863 -0.54377903## [ reached getOption("max.print") -- omitted 3000 entries ]Transform
Let's look at this again on the probability scale using brms::inv_logit_scaled() to make the transformation.
ggplot(did12, aes(inv_logit_scaled(pred))) + geom_histogram(fill = "#61adff", color = "white") + geom_vline(aes(xintercept = inv_logit_scaled(did_median)), data = did12_medians, color = "magenta", size = 2) + facet_wrap(~did, ncol = 1)Exponentiation
We can exponentiate the log-odds to get normal odds
These are fairly interpretable (especially when greater than 1)
# probabilityinv_logit_scaled(did12_medians$did_median)## [1] 0.03741181 0.59435448# oddsexp(did12_medians$did_median)## [1] 0.03886586 1.46520658# odds of the differenceexp(diff(did12_medians$did_median))## [1] 37.69907Confidence in difference?
Everything is a distribution
Just compute the difference in these distributions, and we get a new distribution, which we can use to summarize our uncertainty
did12_wider <- tibble( did1 = pred_did1, did2 = pred_did2) %>% mutate(diff = did2 - did1)did12_wider## # A tibble: 4,000 x 3## did1 did2 diff## <dbl> <dbl> <dbl>## 1 -2.440044 0.7427359 3.18278 ## 2 -8.672287 -0.2324662 8.439821## 3 -4.031558 0.2963330 4.327891## 4 -2.585353 0.8626371 3.44799 ## 5 -3.778682 0.2879902 4.066672## 6 -2.610537 0.2724074 2.882944## 7 -3.817797 0.7147088 4.532506## 8 -3.130512 0.6158916 3.746404## 9 -3.207430 0.4442302 3.65166 ## 10 -5.657550 0.2998222 5.957372## # … with 3,990 more rowsDirectionality
Let's say we want to simplify the question to directionality.
Is there a greater chance of remission for did 2 than 1?
table(did12_wider$diff > 0) / 4000## ## TRUE ## 1The distributions are not overlapping at all - therefore, we are as certain as we can be that the odds of remission are higher with did 2 than 1.
One more quick example
Let's do the same thing, but comparing did 2 and 3.
did3 <- filter(dids, did == 3)pred_did3 <- pop_level + did3$r_diddid23 <- did12_wider %>% select(-did1, -diff) %>% mutate(did3 = pred_did3, diff = did3 - did2)did23## # A tibble: 4,000 x 3## did2 did3 diff## <dbl> <dbl> <dbl>## 1 0.7427359 2.316826 1.57409 ## 2 -0.2324662 1.038893 1.271359## 3 0.2963330 1.632892 1.336559## 4 0.8626371 1.552087 0.68945 ## 5 0.2879902 1.575678 1.287688## 6 0.2724074 1.483903 1.211496## 7 0.7147088 1.749563 1.034854## 8 0.6158916 1.229078 0.613186## 9 0.4442302 1.474110 1.02988 ## 10 0.2998222 1.705350 1.405528## # … with 3,990 more rowsPlot data
pd23 <- did23 %>% pivot_longer(did2:diff, names_to = "Distribution", values_to = "Log-Odds")pd23## # A tibble: 12,000 x 2## Distribution `Log-Odds`## <chr> <dbl>## 1 did2 0.7427359## 2 did3 2.316826 ## 3 diff 1.57409 ## 4 did2 -0.2324662## 5 did3 1.038893 ## 6 diff 1.271359 ## 7 did2 0.2963330## 8 did3 1.632892 ## 9 diff 1.336559 ## 10 did2 0.8626371## # … with 11,990 more rowsMissing data on the IVs
Much more problematic, no matter the model or application
Remove all cases with any missingness on any IV?
Limits your sample size
Might (probably?) introduces new sources of bias
Impute?
- Often ethical challenges here - do you really want to impute somebody's gender?
Missing data on the IVs
Much more problematic, no matter the model or application
Remove all cases with any missingness on any IV?
Limits your sample size
Might (probably?) introduces new sources of bias
Impute?
- Often ethical challenges here - do you really want to impute somebody's gender?

Solution?
There really isn't a great one. Be clear about the decisions you do make.
If you do choose imputation, use multiple imputation
- This will allow you to have uncertainty in your imputation
The purpose is to get unbiased population estimates for your parameters (not make inferences about an individual for whom data were imputed)
Missing IDs
In multilevel models, you always have IDs linking the data to the higher levels
If you are missing these IDs, I'm not really sure what to tell you
This is particularly common with longitudinal data (e.g., missing prior school IDs)
In rare cases, you can make assumptions and impute, but those are few and far between, in my experience, and the assumptions are still pretty dangerous
MI for BMI
mi_nhanes$imp$bmi## 1 2 3 4 5## 1 30.1 27.2 33.2 29.6 30.1## 3 30.1 30.1 29.6 26.3 30.1## 4 21.7 22.5 27.4 22.5 25.5## 6 24.9 24.9 21.7 21.7 25.5## 10 28.7 27.4 27.2 20.4 27.4## 11 30.1 28.7 22.0 28.7 35.3## 12 27.5 22.0 22.5 27.4 28.7## 16 35.3 27.2 27.2 26.3 30.1## 21 29.6 22.0 22.0 26.3 33.2Missing formula
We specify a model for each column that has missingness
We have missing data in bmi and chl (not age).
bmi is our outcome, and it will be modeled by age and the complete (missing data imputed) chl variable, as well as their interaction
The missing data in chl will be imputed via a model with age as its predictor!
We're basically fitting two models at once.
In code
The | mi() part says to include missing data, while `
bayes_impute_formula <- bf(bmi | mi() ~ age * mi(chl)) + # base model bf(chl | mi() ~ age) + # model for chl missingness set_rescor(FALSE) # we don't estimate the residual correlationFit
m_onfly <- brm(bayes_impute_formula, data = nhanes)Comparison
Multiple imputation before modeling
conditional_effects(m_mice, "age:chl", resp = "bmi")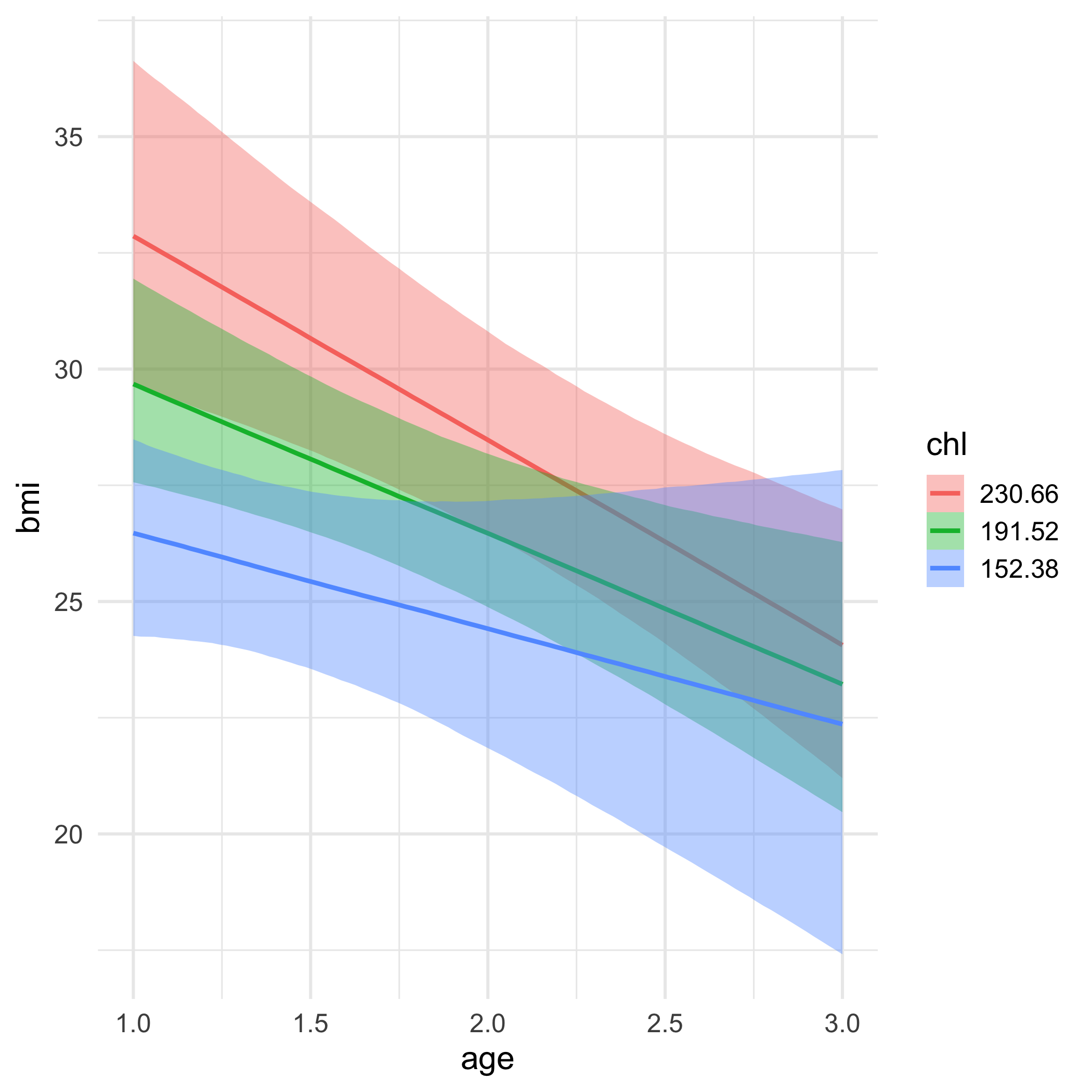
Comparison
imputation during modeling
conditional_effects(m_onfly, "age:chl", resp = "bmi")